Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments
- PMID: 10831421
- PMCID: PMC110551
- DOI: 10.1128/AEM.66.6.2430-2437.2000
Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments
Abstract
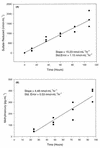
Differences in methylmercury (CH(3)Hg) production normalized to the sulfate reduction rate (SRR) in various species of sulfate-reducing bacteria (SRB) were quantified in pure cultures and in marine sediment slurries in order to determine if SRB strains which differ phylogenetically methylate mercury (Hg) at similar rates. Cultures representing five genera of the SRB (Desulfovibrio desulfuricans, Desulfobulbus propionicus, Desulfococcus multivorans, Desulfobacter sp. strain BG-8, and Desulfobacterium sp. strain BG-33) were grown in a strictly anoxic, minimal medium that received a dose of inorganic Hg 120 h after inoculation. The mercury methylation rates (MMR) normalized per cell were up to 3 orders of magnitude higher in pure cultures of members of SRB groups capable of acetate utilization (e.g., the family Desulfobacteriaceae) than in pure cultures of members of groups that are not able to use acetate (e.g., the family Desulfovibrionaceae). Little or no Hg methylation was observed in cultures of Desulfobacterium or Desulfovibrio strains in the absence of sulfate, indicating that Hg methylation was coupled to respiration in these strains. Mercury methylation, sulfate reduction, and the identities of sulfate-reducing bacteria in marine sediment slurries were also studied. Sulfate-reducing consortia were identified by using group-specific oligonucleotide probes that targeted the 16S rRNA molecule. Acetate-amended slurries, which were dominated by members of the Desulfobacterium and Desulfobacter groups, exhibited a pronounced ability to methylate Hg when the MMR were normalized to the SRR, while lactate-amended and control slurries had normalized MMR that were not statistically different. Collectively, the results of pure-culture and amended-sediment experiments suggest that members of the family Desulfobacteriaceae have a greater potential to methylate Hg than members of the family Desulfovibrionaceae have when the MMR are normalized to the SRR. Hg methylation potential may be related to genetic composition and/or carbon metabolism in the SRB. Furthermore, we found that in marine sediments that are rich in organic matter and dissolved sulfide rapid CH(3)Hg accumulation is coupled to rapid sulfate reduction. The observations described above have broad implications for understanding the control of CH(3)Hg formation and for developing remediation strategies for Hg-contaminated sediments.
Figures





Similar articles
-
Effects of sulfate reducing bacteria and sulfate concentrations on mercury methylation in freshwater sediments.Sci Total Environ. 2012 May 1;424:331-6. doi: 10.1016/j.scitotenv.2011.09.042. Epub 2012 Mar 22. Sci Total Environ. 2012. PMID: 22444059
-
Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria.Appl Environ Microbiol. 2003 Sep;69(9):5414-22. doi: 10.1128/AEM.69.9.5414-5422.2003. Appl Environ Microbiol. 2003. PMID: 12957930 Free PMC article.
-
Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria.Environ Sci Technol. 2008 Jan 1;42(1):93-9. doi: 10.1021/es0705644. Environ Sci Technol. 2008. PMID: 18350881
-
The distribution and activity of sulphate reducing bacteria in estuarine and coastal marine sediments.Antonie Van Leeuwenhoek. 2002 Aug;81(1-4):181-7. doi: 10.1023/a:1020550215012. Antonie Van Leeuwenhoek. 2002. PMID: 12448716 Review.
-
Sulfate-reducing bacteria and sulfur-oxidizing bacteria interactions at redox interfaces: Implications for mercury methylation.Environ Res. 2025 Aug 7;285(Pt 3):122553. doi: 10.1016/j.envres.2025.122553. Online ahead of print. Environ Res. 2025. PMID: 40782964 Review.
Cited by
-
Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species.Appl Environ Microbiol. 2012 Oct;78(20):7337-46. doi: 10.1128/AEM.01792-12. Epub 2012 Aug 10. Appl Environ Microbiol. 2012. PMID: 22885751 Free PMC article.
-
Two-component signal transduction systems of Desulfovibrio vulgaris: structural and phylogenetic analysis and deduction of putative cognate pairs.J Mol Evol. 2006 Apr;62(4):473-87. doi: 10.1007/s00239-005-0116-1. Epub 2006 Mar 17. J Mol Evol. 2006. PMID: 16547644
-
Relationships between bacterial energetic metabolism, mercury methylation potential, and hgcA/hgcB gene expression in Desulfovibrio dechloroacetivorans BerOc1.Environ Sci Pollut Res Int. 2015 Sep;22(18):13764-71. doi: 10.1007/s11356-015-4273-5. Epub 2015 Mar 14. Environ Sci Pollut Res Int. 2015. PMID: 25772867
-
Neurotoxicity of organomercurial compounds.Neurotox Res. 2003;5(4):283-305. doi: 10.1007/BF03033386. Neurotox Res. 2003. PMID: 12835120 Review.
-
Mercury analysis of acid- and alkaline-reduced biological samples: identification of meta-cinnabar as the major biotransformed compound in algae.Appl Environ Microbiol. 2006 Jan;72(1):361-7. doi: 10.1128/AEM.72.1.361-367.2006. Appl Environ Microbiol. 2006. PMID: 16391065 Free PMC article.
References
-
- Andersson I, Parkman H, Jernelov A. The role of sediments as sink or source for environmental contaminants: a case study of Hg and chlorinated organic compounds. Limnologica. 1990;20:347–359.
-
- Benoit J M, Gilmour C G, Mason R P, Heyes A. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ Sci Technol. 1999;33:951–957.
-
- Benoit J M, Mason R P, Gilmour C G. Estimation of mercury-sulfide speciation in sediment pore waters using octanol-water partitioning and implication for the availability to methylating bacteria. Environ Toxicol Chem. 1999;18:2138–2141. - PubMed
-
- Blum J E, Bartha R. Effects of salinity on methylation of Hg. Bull Environ Contam Toxicol. 1980;25:404–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

