Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster
- PMID: 10831439
- PMCID: PMC110582
- DOI: 10.1128/AEM.66.6.2565-2571.2000
Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster
Abstract
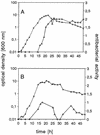
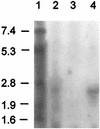
The biosynthetic gene cluster (12.3 kb) of mersacidin, a lanthionine-containing antimicrobial peptide, is located on the chromosome of the producer, Bacillus sp. strain HIL Y-85,54728 in a region that corresponds to 348 degrees on the chromosome of Bacillus subtilis 168. It consists of 10 open reading frames and contains, in addition to the previously described mersacidin structural gene mrsA (G. Bierbaum, H. Brötz, K.-P. Koller, and H.-G. Sahl, FEMS Microbiol. Lett. 127:121-126, 1995), two genes, mrsM and mrsD, coding for enzymes involved in posttranslational modification of the prepeptide; one gene, mrsT, coding for a transporter with an associated protease domain; and three genes, mrsF, mrsG, and mrsE, encoding a group B ABC transporter that could be involved in producer self-protection. Additionally, three regulatory genes are part of the gene cluster, i.e., mrsR2 and mrsK2, which encode a two-component regulatory system which seems to be necessary for the transcription of the mrsFGE operon, and mrsR1, which encodes a protein with similarity to response regulators. Transcription of mrsA sets in at early stationary phase (between 8 and 16 h of culture).
Figures




Similar articles
-
Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity.Appl Environ Microbiol. 2002 Jan;68(1):106-13. doi: 10.1128/AEM.68.1.106-113.2002. Appl Environ Microbiol. 2002. PMID: 11772616 Free PMC article.
-
The lantibiotic mersacidin is an autoinducing peptide.Appl Environ Microbiol. 2006 Nov;72(11):7270-7. doi: 10.1128/AEM.00723-06. Epub 2006 Sep 15. Appl Environ Microbiol. 2006. PMID: 16980420 Free PMC article.
-
Construction of an expression system for site-directed mutagenesis of the lantibiotic mersacidin.Appl Environ Microbiol. 2003 Jul;69(7):3777-83. doi: 10.1128/AEM.69.7.3777-3783.2003. Appl Environ Microbiol. 2003. PMID: 12839744 Free PMC article.
-
Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics.Antonie Van Leeuwenhoek. 1996 Feb;69(2):109-17. doi: 10.1007/BF00399416. Antonie Van Leeuwenhoek. 1996. PMID: 8775971 Review.
-
Comparison of lantibiotic gene clusters and encoded proteins.Antonie Van Leeuwenhoek. 1996 Feb;69(2):171-84. doi: 10.1007/BF00399422. Antonie Van Leeuwenhoek. 1996. PMID: 8775977 Review.
Cited by
-
Overproduction of wild-type and bioengineered derivatives of the lantibiotic lacticin 3147.Appl Environ Microbiol. 2006 Jun;72(6):4492-6. doi: 10.1128/AEM.02543-05. Appl Environ Microbiol. 2006. PMID: 16751576 Free PMC article.
-
Characterization, distribution, and expression of novel genes among eight clinical isolates of Streptococcus pneumoniae.Infect Immun. 2006 Jan;74(1):321-30. doi: 10.1128/IAI.74.1.321-330.2006. Infect Immun. 2006. PMID: 16368987 Free PMC article.
-
Characterization of amylolysin, a novel lantibiotic from Bacillus amyloliquefaciens GA1.PLoS One. 2013 Dec 9;8(12):e83037. doi: 10.1371/journal.pone.0083037. eCollection 2013. PLoS One. 2013. PMID: 24349428 Free PMC article.
-
Lactolisterin BU, a Novel Class II Broad-Spectrum Bacteriocin from Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4.Appl Environ Microbiol. 2017 Oct 17;83(21):e01519-17. doi: 10.1128/AEM.01519-17. Print 2017 Nov 1. Appl Environ Microbiol. 2017. PMID: 28842543 Free PMC article.
-
Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering.Microb Biotechnol. 2019 Sep;12(5):828-844. doi: 10.1111/1751-7915.13398. Epub 2019 Mar 4. Microb Biotechnol. 2019. PMID: 30834674 Free PMC article. Review.
References
-
- Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K-D, Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. - PubMed
-
- Bierbaum G, Brötz H, Koller K-P, Sahl H-G. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol Lett. 1995;127:121–126. - PubMed
-
- Bisschop A, Konings W N. Reconstitution of reduced nicotinamide adenine dinucleotide oxidase activity with menadione in membrane vesicles from the menaquinone-deficient Bacillus subtilis AroD. Eur J Biochem. 1976;67:357–365. - PubMed
-
- Booth M C, Bogie C P, Sahl H-G, Siezen R J, Hatter K L, Gilmore M S. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol. 1996;21:1175–1184. - PubMed
-
- Brötz H, Bierbaum G, Reynolds P E, Sahl H-G. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur J Biochem. 1997;246:193–199. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

