Use of repetitive DNA sequences and the PCR To differentiate Escherichia coli isolates from human and animal sources
- PMID: 10831440
- PMCID: PMC110583
- DOI: 10.1128/AEM.66.6.2572-2577.2000
Use of repetitive DNA sequences and the PCR To differentiate Escherichia coli isolates from human and animal sources
Abstract
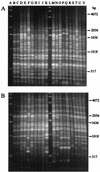
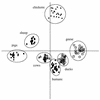
The rep-PCR DNA fingerprint technique, which uses repetitive intergenic DNA sequences, was investigated as a way to differentiate between human and animal sources of fecal pollution. BOX and REP primers were used to generate DNA fingerprints from Escherichia coli strains isolated from human and animal sources (geese, ducks, cows, pigs, chickens, and sheep). Our initial studies revealed that the DNA fingerprints obtained with the BOX primer were more effective for grouping E. coli strains than the DNA fingerprints obtained with REP primers. The BOX primer DNA fingerprints of 154 E. coli isolates were analyzed by using the Jaccard band-matching algorithm. Jackknife analysis of the resulting similarity coefficients revealed that 100% of the chicken and cow isolates and between 78 and 90% of the human, goose, duck, pig, and sheep isolates were assigned to the correct source groups. A dendrogram constructed by using Jaccard similarity coefficients almost completely separated the human isolates from the nonhuman isolates. Multivariate analysis of variance, a form of discriminant analysis, successfully differentiated the isolates and placed them in the appropriate source groups. Taken together, our results indicate that rep-PCR performed with the BOX A1R primer may be a useful and effective tool for rapidly determining sources of fecal pollution.
Figures

References
-
- Bouzar H, Jones J B, Stall R E, Louws F J, Schneider M, Rademaker J L W, de Bruijn F J, Jackson L E. Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: evidence for common lineages within and between countries. Phytopathology. 1999;89:328–335. - PubMed
-
- de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. - PMC - PubMed
-
- Devriese L A, Van De Kerckhove A, Kilpper-Bälz R, Schleifer K H. Characterization and identification of Enterococcus species isolated from the intestines of animals. Int J Syst Bacteriol. 1987;37:257–259.
-
- Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. - PubMed
-
- Feachem R. An improved role for faecal coliform to faecal streptococci ratios in the differentiation between human and nonhuman pollution sources. Water Res. 1975;9:689–690.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

