Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana
- PMID: 10835031
- PMCID: PMC86843
- DOI: 10.1128/JCM.38.6.2453-2456.2000
Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana
Abstract
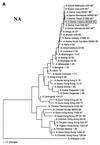
An H1N2 influenza virus was isolated from a pig during an outbreak of respiratory disease and abortion on an Indiana farm in November 1999. Results of phylogenetic analyses indicate that this virus is a reassortant between a recent classical H1 swine virus and the reassortant H3N2 viruses that have emerged among American pigs since 1998.
Figures



References
-
- Brown I H, Chakraverty P, Harris P A, Alexander D J. Disease outbreaks in pigs in Great Britain due to an influenza A virus of H1N2 subtype. Vet Rec. 1995;136:328–329. - PubMed
-
- Brown I H, Harris P A, McCauley J W, Alexander D J. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79:2947–2955. - PubMed
-
- Campitelli L, Donatelli I, Foni E, Castrucci M R, Fabiani C, Kawaoka Y, Krauss S, Webster R G. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. - PubMed
-
- Castrucci M R, Campitelli L, Ruggieri A, Barigazzi G, Sidoli L, Daniels R, Oxford J S, Donatelli I. Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J Gen Virol. 1994;75:371–379. - PubMed
-
- Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza viruses in Italian pigs. Virology. 1993;193:503–506. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

