One protein from two open reading frames: mechanism of a 50 nt translational bypass
- PMID: 10835364
- PMCID: PMC212773
- DOI: 10.1093/emboj/19.11.2671
One protein from two open reading frames: mechanism of a 50 nt translational bypass
Abstract
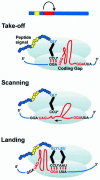
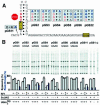
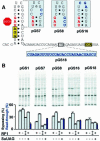
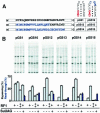
Translating ribosomes bypass a 50 nt coding gap in order to fuse the information found in the two open reading frames (ORFs) of bacteriophage T4 gene 60. This study investigates the underlying mechanism by focusing on the competition between initiation of bypassing and termination at the end of the first ORF. While nearly all ribosomes initiate bypassing, no more than 50% resume translation in the second ORF. Two previously described cis-acting stimulatory signals are critical for favoring initiation of bypassing over termination. Genetic analysis of these signals supports a working model in which the first (a stem-loop structure at the junction between the first ORF and the coding gap) interferes with decoding in the A-site, and the second (a stretch of amino acids in the nascent peptide encoded by the first ORF) destabilizes peptidyl-tRNA-mRNA pairing.
Figures

References
-
- Adamski F.M., Atkins,J.F. and Gesteland,R.F. (1996) Ribosomal protein L9 interactions with 23S rRNA: the use of a translational bypass assay to study the effect of amino acid substitutions. J. Mol. Biol., 261, 357–371. - PubMed
-
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.
-
- Bossi L. and Roth,J.R. (1980) The influence of codon context on genetic code translation. Nature, 286, 123–127. - PubMed
-
- Brimacombe R., Gornicki,P., Greuer,B., Mitchell,P., Osswald,M., Rinke-Appel,J., Schuler,D. and Stade,K. (1990) The three-dimensional structure and function of Escherichia coli ribosomal RNA, as studied by cross-linking techniques. Biochim. Biophys. Acta, 1050, 8–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

