Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution
- PMID: 10835368
- PMCID: PMC212750
- DOI: 10.1093/emboj/19.11.2710
Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution
Abstract
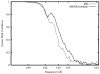
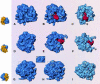
Using a sordarin derivative, an antifungal drug, it was possible to determine the structure of a eukaryotic ribosome small middle dotEF2 complex at 17.5 A resolution by three-dimensional (3D) cryo-electron microscopy. EF2 is directly visible in the 3D map and the overall arrangement of the complex from Saccharomyces cerevisiae corresponds to that previously seen in Escherichia coli. However, pronounced differences were found in two prominent regions. First, in the yeast system the interaction between the elongation factor and the stalk region of the large subunit is much more extensive. Secondly, domain IV of EF2 contains additional mass that appears to interact with the head of the 40S subunit and the region of the main bridge of the 60S subunit. The shape and position of domain IV of EF2 suggest that it might interact directly with P-site-bound tRNA.
Figures



References
-
- Abel K. and Jurnak,F. (1996) A complex profile of protein elongation: translating chemical energy into molecular movement. Structure, 4, 229–238. - PubMed
-
- Agrawal R.K., Heagle,A.B., Penczek,P., Grassucci,R.A. and Frank,J. (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Struct. Biol., 6, 643–647. - PubMed
-
- Agrawal R.K., Heagle,A.B. and Frank,J. (2000) Studies of elongation factor G-dependent tRNA translocation by three-dimensional cryo-electron microscopy. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 53–62.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

