N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation
- PMID: 10837031
- PMCID: PMC316670
N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation
Abstract
Members of the myc family of cellular oncogenes have been implicated as transcriptional regulators in pathways that govern cellular proliferation and death. In addition, N-myc and c-myc are essential for completion of murine embryonic development. However, the basis for the evolutionary conservation of myc gene family has remained unclear. To elucidate this issue, we have generated mice in which the endogenous c-myc coding sequences have been replaced with N-myc coding sequences. Strikingly, mice homozygous for this replacement mutation can survive into adulthood and reproduce. Moreover, when expressed from the c-myc locus, N-myc is similarly regulated and functionally complementary to c-myc in the context of various cellular growth and differentiation processes. Therefore, the myc gene family must have evolved, to a large extent, to facilitate differential patterns of expression.
Figures

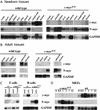
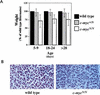
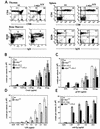
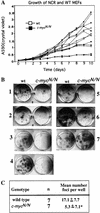
References
-
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. - PubMed
-
- Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. - PubMed
-
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes & Dev. 1992;6:2248–2257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
