Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro
- PMID: 10840064
- PMCID: PMC16534
- DOI: 10.1073/pnas.130189697
Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro
Abstract
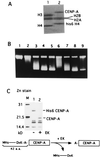
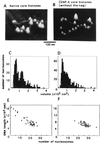
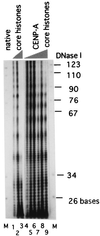
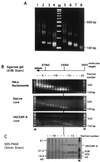
Centromere protein A (CENP-A) is a variant of histone H3 with more than 60% sequence identity at the C-terminal histone fold domain. CENP-A specifically locates to active centromeres of animal chromosomes and therefore is believed to be a component of the specialized centromeric nucleosomes on which the kinetochores are assembled. Here we report that CENP-A, highly purified from HeLa cells, can indeed replace histone H3 in a nucleosome reconstitution system mediated by nucleosome assembly protein-1 (NAP-1). The structure of the nucleosomes reconstituted with recombinant CENP-A, histones H2A, H2B, and H4, and closed circular DNAs had the following properties. By atomic force microscopy, "beads on a string" images were obtained that were similar to those obtained with nucleosomes reconstituted with four standard histones. DNA ladders with repeats of approximately 10 bp were produced by DNase I digestion, indicating that the DNA was wrapped round the protein complex. Mononucleosomes isolated by glycerol gradient sedimentation had a relative molecular mass of approximately 200 kDa and were composed of 120-150 bp of DNA and equimolar amounts of CENP-A, and histones H4, H2A, and H2B. Thus, we conclude that CENP-A forms an octameric complex with histones H4, H2A, and H2B in the presence of DNA.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

