A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family
- PMID: 10841563
- PMCID: PMC18692
- DOI: 10.1073/pnas.97.12.6658
A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family
Abstract
The yeast Sir2 protein, required for transcriptional silencing, has an NAD(+)-dependent histone deacetylase (HDA) activity. Yeast extracts contain a NAD(+)-dependent HDA activity that is eliminated in a yeast strain from which SIR2 and its four homologs have been deleted. This HDA activity is also displayed by purified yeast Sir2p and homologous Archaeal, eubacterial, and human proteins, and depends completely on NAD(+) in all species tested. The yeast NPT1 gene, encoding an important NAD(+) synthesis enzyme, is required for rDNA and telomeric silencing and contributes to silencing of the HM loci. Null mutants in this gene have significantly reduced intracellular NAD(+) concentrations and have phenotypes similar to sir2 null mutants. Surprisingly, yeast from which all five SIR2 homologs have been deleted have relatively normal bulk histone acetylation levels. The evolutionary conservation of this regulated activity suggests that the Sir2 protein family represents a set of effector proteins in an evolutionarily conserved signal transduction pathway that monitors cellular energy and redox states.
Figures

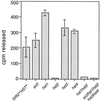
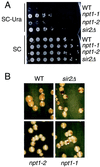

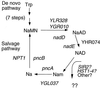

References
-
- Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. - PubMed
-
- Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. - PubMed
-
- Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. - PubMed
-
- Bryk M, Banerjee M, Murphy M, Knudsen K E, Garfinkel D J, Curcio M J. Genes Dev. 1997;11:255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

