Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis
- PMID: 10848492
- PMCID: PMC1287103
- DOI: 10.1086/302995
Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis
Abstract
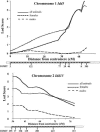
Insulin-dependent diabetes mellitus (IDDM) is a polygenic disease caused by progressive autoimmune infiltration (insulitis) of the pancreatic islets of Langerhan, culminating in the destruction of insulin-producing beta cells. Genome scans of families with diabetes suggest that multiple loci make incremental contributions to disease susceptibility. However, only the IDDM1 locus is well characterized, at a molecular and functional level, as alleleic variants of the major histocompatibility complex (MHC) class II HLA-DQB1, DRB1, and DPB1 genes that mediate antigen presentation to T cells. In the nonobese diabetic (NOD) mouse model, the Idd1 locus was shown to be the orthologous MHC gene I-Ab. Inheritance of susceptibility alleles at IDDM1/Idd1 is insufficient for disease development in humans and NOD mice. However, the identities and functions of the remaining diabetes loci (Idd2-Idd19 in NOD mice) are largely undefined. A crucial limitation in previous genetic linkage studies of this disease has been reliance on a single complex phenotype-diabetes that displays low penetrance and is of limited utility for high-resolution genetic mapping. Using the NOD model, we have identified an early step in diabetes pathogenesis that behaves as a highly penetrant trait. We report that NOD-derived alleles at both the Idd5 and Idd13 loci regulate a T lymphocyte-dependent progression from a benign to a destructive stage of insulitis. Human chromosomal regions orthologous to the Idd5 and -13 intervals are also linked to diabetes risk, suggesting that conserved genes encoded at these loci are central regulators of disease pathogenesis. These data are the first to reveal a role for individual non-MHC Idd loci in a specific, critical step in diabetes pathogenesis-T cell recruitment to islet lesions driving destructive inflammation. Importantly, identification of intermediate phenotypes in complex disease pathogenesis provides the tools required to progress toward gene identification at these loci.
Figures




References
Electronic-Database Information
-
- Genome Center Software Information and Documentation, http://waldo.wi.mit.edu/genome_software
-
- Jackson Laboratory, http://www.jax.org
-
- NCBI Human/Mouse Homology Relationships, http://www.ncbi.nlm.gov/Homology
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for IDDM1 [MIM 222100]
-
- Research Genetics, http://www.resgen.com
References
-
- Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, et al (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 9:284–292 - PubMed
-
- Bennett ST, Todd JA (1996) Human type 1 diabetes and the insulin gene: principles of mapping polygenes. Annu Rev Genet 30:343–370 - PubMed
-
- Bonnevie-Nielsen V, Skovgaard LT, Lernmark A (1983) β-Cell function relative to islet volume and hormone content in the isolated perfused mouse pancreas. Endocrinology 112:1049–1056 - PubMed
-
- Caillat-Zucman S, Daniel S, Djilali-Saiah I, Timsit J, Garchon HJ, Boitard C, Bach JF (1995) Family study of linkage disequilibrium between TAP2 transporter and HLA class II genes: absence of TAP2 contribution to association with insulin-dependent diabetes mellitus. Hum Immunol 44:80–87 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

