PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex
- PMID: 10848580
- PMCID: PMC85837
- DOI: 10.1128/MCB.20.13.4532-4542.2000
PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex
Abstract
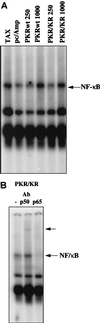
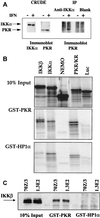
The interferon (IFN)-induced double-stranded RNA-activated protein kinase PKR mediates inhibition of protein synthesis through phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2alpha) and is also involved in the induction of the IFN gene through the activation of the transcription factor NF-kappaB. NF-kappaB is retained in the cytoplasm through binding to its inhibitor IkappaBalpha. The critical step in NF-kappaB activation is the phosphorylation of IkappaBalpha by the IkappaB kinase (IKK) complex. This activity releases NF-kappaB from IkappaBalpha and allows its translocation to the nucleus. Here, we have studied the ability of PKR to activate NF-kappaB in a reporter assay and have shown for the first time that two catalytically inactive PKR mutants, PKR/KR296 and a deletion mutant (PKR/Del42) which lacks the potential eIF2alpha-binding domain, can also activate NF-kappaB. This result indicated that NF-kappaB activation by PKR does not require its kinase activity and that it is independent of the PKR-eIF2alpha relationship. Transfection of either wild-type PKR or catalytically inactive PKR in PKR(0/0) mouse embryo fibroblasts resulted in the activation of the IKK complex. By using a glutathione S-transferase pull-down assay, we showed that PKR interacts with the IKKbeta subunit of the IKK complex. This interaction apparently does not require the integrity of the IKK complex, as it was found to occur with extracts from cells deficient in the NF-kappaB essential modulator, one of the components of the IKK complex. Therefore, our results reveal a novel pathway by which PKR can modulate the NF-kappaB signaling pathway without using its kinase activity.
Figures


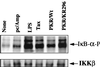
References
-
- Baeuerle P, Baichwal V. NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv Immunol. 1997;65:111–136. - PubMed
-
- Baldwin A. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. - PubMed
-
- Berlanga J J, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur J Biochem. 1999;265:754–762. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
