Role of DBP in the circadian oscillatory mechanism
- PMID: 10848603
- PMCID: PMC85912
- DOI: 10.1128/MCB.20.13.4773-4781.2000
Role of DBP in the circadian oscillatory mechanism
Abstract
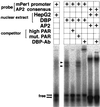
Transcript levels of DBP, a member of the PAR leucine zipper transcription factor family, exhibit a robust rhythm in suprachiasmatic nuclei, the mammalian circadian center. Here we report that DBP is able to activate the promoter of a putative clock oscillating gene, mPer1, by directly binding to the mPer1 promoter. The mPer1 promoter is cooperatively activated by DBP and CLOCK-BMAL1. On the other hand, dbp transcription is activated by CLOCK-BMAL1 through E-boxes and inhibited by the mPER and mCRY proteins, as is the case for mPer1. Thus, a clock-controlled dbp gene may play an important role in central clock oscillation.
Figures






References
-
- Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. - PubMed
-
- Blau J, Young M W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. - PubMed
-
- Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbach M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. - PubMed
-
- Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
