Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable
- PMID: 10848611
- PMCID: PMC85937
- DOI: 10.1128/MCB.20.13.4859-4869.2000
Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable
Abstract
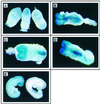
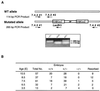
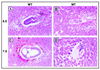
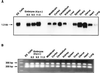
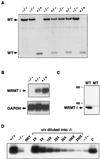
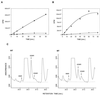
Protein arginine N-methyltransferases have been implicated in a variety of processes, including cell proliferation, signal transduction, and protein trafficking. In this study, we have characterized essentially a null mutation induced by insertion of the U3betaGeo gene trap retrovirus into the second intron of the mouse protein arginine N-methyltransferase 1 gene (Prmt1). cDNAs encoding two forms of Prmt1 were characterized, and the predicted protein sequences were found to be highly conserved among vertebrates. Expression of the Prmt1-betageo fusion gene was greatest along the midline of the neural plate and in the forming head fold from embryonic day 7.5 (E7.5) to E8.5 and in the developing central nervous system from E8.5 to E13.5. Homozygous mutant embryos failed to develop beyond E6.5, a phenotype consistent with a fundamental role in cellular metabolism. However, Prmt1 was not required for cell viability, as the protein was not detected in embryonic stem (ES) cell lines established from mutant blastocysts. Low levels of Prmt1 transcripts (approximately 1% of the wild-type level) were detected as assessed by a quantitative reverse transcription-PCR assay. Total levels of arginine N-methyltransferase activity and asymmetric N(G), N(G)-dimethylarginine were reduced by 85 and 54%, respectively, while levels of hypomethylated substrates were increased 15-fold. Prmt1 appears to be a major type I enzyme in ES cells, and in wild-type cells, most substrates of the enzyme appear to be maintained in a fully methylated state.
Figures



References
-
- Byvoet P, Shepherd G R, Hardin J M, Noland B J. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148:558–567. - PubMed
-
- Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
