Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon
- PMID: 10848614
- PMCID: PMC85940
- DOI: 10.1128/MCB.20.13.4888-4899.2000
Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon
Abstract
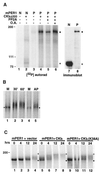
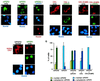
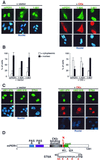
The molecular oscillator that keeps circadian time is generated by a negative feedback loop. Nuclear entry of circadian regulatory proteins that inhibit transcription from E-box-containing promoters appears to be a critical component of this loop in both Drosophila and mammals. The Drosophila double-time gene product, a casein kinase I epsilon (CKIepsilon) homolog, has been reported to interact with dPER and regulate circadian cycle length. We find that mammalian CKIepsilon binds to and phosphorylates the murine circadian regulator mPER1. Unlike both dPER and mPER2, mPER1 expressed alone in HEK 293 cells is predominantly a nuclear protein. Two distinct mechanisms appear to retard mPER1 nuclear entry. First, coexpression of mPER2 leads to mPER1-mPER2 heterodimer formation and cytoplasmic colocalization. Second, coexpression of CKIepsilon leads to masking of the mPER1 nuclear localization signal and phosphorylation-dependent cytoplasmic retention of both proteins. CKIepsilon may regulate mammalian circadian rhythm by controlling the rate at which mPER1 enters the nucleus.
Figures




Similar articles
-
Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells.Mol Cell Biol. 2002 Mar;22(6):1693-703. doi: 10.1128/MCB.22.6.1693-1703.2002. Mol Cell Biol. 2002. PMID: 11865049 Free PMC article.
-
Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock.Mol Cell Biol. 2004 Jan;24(2):584-94. doi: 10.1128/MCB.24.2.584-594.2004. Mol Cell Biol. 2004. PMID: 14701732 Free PMC article.
-
A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus.Genes Cells. 1998 Mar;3(3):167-76. doi: 10.1046/j.1365-2443.1998.00178.x. Genes Cells. 1998. PMID: 9619629
-
Casein kinase I: another cog in the circadian clockworks.Chronobiol Int. 2001 May;18(3):389-98. doi: 10.1081/cbi-100103963. Chronobiol Int. 2001. PMID: 11475410 Review.
-
Role of phosphorylation in the mammalian circadian clock.Cold Spring Harb Symp Quant Biol. 2007;72:167-76. doi: 10.1101/sqb.2007.72.036. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419274 Review.
Cited by
-
Role of type II protein arginine methyltransferase 5 in the regulation of Circadian Per1 gene.PLoS One. 2012;7(10):e48152. doi: 10.1371/journal.pone.0048152. Epub 2012 Oct 25. PLoS One. 2012. PMID: 23133559 Free PMC article.
-
FRQ-CK1 interaction determines the period of circadian rhythms in Neurospora.Nat Commun. 2019 Sep 25;10(1):4352. doi: 10.1038/s41467-019-12239-w. Nat Commun. 2019. PMID: 31554810 Free PMC article.
-
An opposite role for tau in circadian rhythms revealed by mathematical modeling.Proc Natl Acad Sci U S A. 2006 Jul 11;103(28):10618-23. doi: 10.1073/pnas.0604511103. Epub 2006 Jul 3. Proc Natl Acad Sci U S A. 2006. PMID: 16818876 Free PMC article.
-
A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1.Mol Cell Biol. 2004 May;24(10):4184-95. doi: 10.1128/MCB.24.10.4184-4195.2004. Mol Cell Biol. 2004. PMID: 15121840 Free PMC article.
-
The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon.J Biol Chem. 2002 May 10;277(19):17248-54. doi: 10.1074/jbc.M111466200. Epub 2002 Mar 1. J Biol Chem. 2002. PMID: 11875063 Free PMC article.
References
-
- Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. - PubMed
-
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. - PubMed
-
- Briggs L J, Stein D, Goltz J, Corrigan V C, Efthymiadis A, Hubner S, Jans D A. The cAMP-dependent protein kinase site (Ser312) enhances dorsal nuclear import through facilitating nuclear localization sequence/importin interaction. J Biol Chem. 1998;273:22745–22752. - PubMed
-
- Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. - PubMed
-
- Cegielska A, Gietzen K F, Rivers A, Virshup D M. Autoinhibition of casein kinase I ɛ (CKIɛ) is relieved by protein phosphatases and limited proteolysis. J Biol Chem. 1998;273:1357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
