Defining a rob regulon in Escherichia coli by using transposon mutagenesis
- PMID: 10850996
- PMCID: PMC94552
- DOI: 10.1128/JB.182.13.3794-3801.2000
Defining a rob regulon in Escherichia coli by using transposon mutagenesis
Abstract
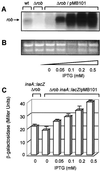
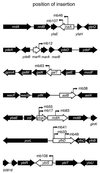
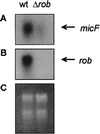
The Rob protein of Escherichia coli is a member of the AraC-XylS family of prokaryotic transcriptional regulators and is expressed constitutively. Deletion of the rob gene increases susceptibility to organic solvents, while overexpression of Rob increases tolerance to organic solvents and resistance to a variety of antibiotics and to the superoxide-generating compound phenazine methosulfate. To determine whether constitutive levels of Rob regulate basal gene expression, we performed a MudJ transposon screen in a rob deletion mutant containing a plasmid that allows for controlled rob gene expression. We identified eight genes and confirmed that seven are transcriptionally activated by normal expression of Rob from the chromosomal rob gene (inaA, marR, aslB, ybaO, mdlA, yfhD, and ybiS). One gene, galT, was repressed by Rob. We also demonstrated by Northern analysis that basal expression of micF is significantly higher in wild-type E. coli than in a rob deletion mutant. Rob binding to the promoter regions of most of these genes was substantiated in electrophoretic mobility shift assays. However, Mu insertions in individual Rob-regulated genes did not affect solvent sensitivity. This phenotype may depend on changes in the expression of several of these Rob-regulated genes or on other genes that were not identified. Rob clearly affects the basal expression of genes with a broad range of functions, including antibiotic resistance, acid adaptation, carbon metabolism, cell wall synthesis, central intermediary metabolism, and transport. The magnitudes of Rob's effects are modest, however, and the protein may thus play a role as a general transcription cofactor.
Figures
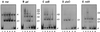
References
-
- Adhya S. The galactose operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1503–1512.
-
- Allikmets R, Gerrard B, Court D, Dean M. Cloning and organization of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene. 1993;136:231–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

