Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus
- PMID: 10852873
- PMCID: PMC101913
- DOI: 10.1128/JB.182.12.3423-3428.2000
Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus
Abstract
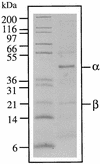
Highly washed membrane preparations from cells of the hyperthermophilic archaeon Pyrococcus furiosus contain high hydrogenase activity (9.4 micromol of H(2) evolved/mg at 80 degrees C) using reduced methyl viologen as the electron donor. The enzyme was solubilized with n-dodecyl-beta-D-maltoside and purified by multistep chromatography in the presence of Triton X-100. The purified preparation contained two major proteins (alpha and beta) in an approximate 1:1 ratio with a minimum molecular mass near 65 kDa and contained approximately 1 Ni and 4 Fe atoms/mol. The reduced enzyme gave rise to an electron paramagnetic resonance signal typical of the so-called Ni-C center of mesophilic NiFe-hydrogenases. Neither highly washed membranes nor the purified enzyme used NAD(P)(H) or P. furiosus ferredoxin as an electron carrier, nor did either catalyze the reduction of elemental sulfur with H(2) as the electron donor. Using N-terminal amino acid sequence information, the genes proposed to encode the alpha and beta subunits were located in the genome database within a putative 14-gene operon (termed mbh). The deduced sequences of the two subunits (Mbh 11 and 12) were distinctly different from those of the four subunits that comprise each of the two cytoplasmic NiFe-hydrogenases of P. furiosus and show that the alpha subunit contains the NiFe-catalytic site. Six of the open reading frames (ORFs) in the operon, including those encoding the alpha and beta subunits, show high sequence similarity (>30% identity) with proteins associated with the membrane-bound NiFe-hydrogenase complexes from Methanosarcina barkeri, Escherichia coli, and Rhodospirillum rubrum. The remaining eight ORFs encode small (<19-kDa) hypothetical proteins. These data suggest that P. furiosus, which was thought to be solely a fermentative organism, may contain a previously unrecognized respiratory system in which H(2) metabolism is coupled to energy conservation.
Figures

References
-
- Adams M W W. The structure and mechanism of iron-hydrogenases. Biochim Biophys Acta. 1990;1020:115–145. - PubMed
-
- Adams M W W, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archaea. Adv Protein Chem. 1996;48:101–180. - PubMed
-
- Adams M W W, Mortenson L E. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. J Biol Chem. 1984;259:7045–7055. - PubMed
-
- Albracht S P J. Intimate relationships of the large and the small subunits of all nickel hydrogenases with two nuclear-encoded subunits of mitochondrial NADH: ubiquinone oxidoreductase. Biochim Biophys Acta. 1993;1144:221–224. - PubMed
-
- Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

