Analysis of the SOS response in Salmonella enterica serovar typhimurium using RNA fingerprinting by arbitrarily primed PCR
- PMID: 10852882
- PMCID: PMC101940
- DOI: 10.1128/JB.182.12.3490-3497.2000
Analysis of the SOS response in Salmonella enterica serovar typhimurium using RNA fingerprinting by arbitrarily primed PCR
Abstract
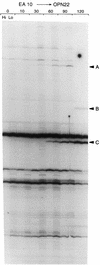
We report an analysis of a sample of the SOS response of Salmonella enterica serovar Typhimurium using the differential display of RNA fingerprinting gels of arbitrarily primed PCR products. The SOS response was induced by the addition of mitomycin C to an exponentially growing culture of serovar Typhimurium, and the RNA population was sampled during the following 2 h. These experiments revealed 21 differentially expressed PCR fragments representing mRNA transcripts. These 21 fragments correspond to 20 distinct genes. All of these transcripts were positively regulated, with the observed induction starting 10 to 120 min after addition of mitomycin C. Fifteen of the 21 transcripts have no homologue in the public sequence data banks and are therefore classified as novel. The remaining six transcripts corresponded to the recE, stpA, sulA, and umuC genes, and to a gene encoding a hypothetical protein in the Escherichia coli lysU-cadA intergenic region; the recE gene was represented twice by nonoverlapping fragments. In order to determine if the induction of these 20 transcripts constitutes part of a classical SOS regulon, we assessed the induction of these genes in a recA mutant. With one exception, the increased expression of these genes in response to mitomycin C was dependent on the presence of a functional recA allele. The exception was fivefold induced in the absence of a functional RecA protein, suggesting another layer of regulation in response to mitomycin C, in addition to the RecA-LexA pathway of SOS induction. Our data reveal several genes belonging to operons known to be directly involved in pathogenesis. In addition, we have found several phage-like sequences, some of which may be landmarks of pathogenicity determinants. On the basis of these observations, we propose that the general use of DNA-damaging agents coupled with differential gene expression analysis may be a useful and easy method for identifying pathogenicity determinants in diverse organisms.
Figures


References
-
- Anderson D G, Kowalczykowski S C. Reconstitution of an SOS response pathway: derepression of transcription in response to DNA breaks. Cell. 1998;95:975–979. - PubMed
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

