Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers
- PMID: 10852939
- PMCID: PMC149095
- DOI: 10.1105/tpc.12.6.949
Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers
Abstract
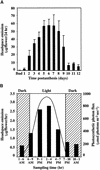
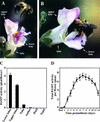
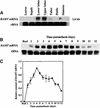
In snapdragon flowers, the volatile ester methyl benzoate is the most abundant scent compound. It is synthesized by and emitted from only the upper and lower lobes of petals, where pollinators (bumblebees) come in contact with the flower. Emission of methyl benzoate occurs in a rhythmic manner, with maximum emission during the day, which correlates with pollinator activity. A novel S-adenosyl-l-methionine:benzoic acid carboxyl methyl transferase (BAMT), the final enzyme in the biosynthesis of methyl benzoate, and its corresponding cDNA have been isolated and characterized. The complete amino acid sequence of the BAMT protein has only low levels of sequence similarity to other previously characterized proteins, including plant O-methyl transferases. During the life span of the flower, the levels of methyl benzoate emission, BAMT activity, BAMT gene expression, and the amounts of BAMT protein and benzoic acid are developmentally and differentially regulated. Linear regression analysis revealed that production of methyl benzoate is regulated by the amount of benzoic acid and the amount of BAMT protein, which in turn is regulated at the transcriptional level.
Figures
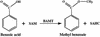




References
-
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. - PubMed
-
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. - PubMed
-
- Coen, E.S., Carpenter, R., and Martin, C. (1986). Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell 47, 285–296. - PubMed
-
- Dobson, H.E.M. (1994). Floral volatiles in insect biology. In Insect–Plant Interactions, Vol. 5, E. Bernays, ed (Boca Raton, FL: CRC Press), pp. 47–81.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

