A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions
- PMID: 10852947
- PMCID: PMC16554
- DOI: 10.1073/pnas.110149297
A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions
Abstract
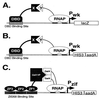
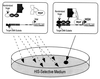
We have developed a bacterial "two-hybrid" system that readily allows selection from libraries larger than 10(8) in size. Our bacterial system may be used to study either protein-DNA or protein-protein interactions, and it offers a number of potentially significant advantages over existing yeast-based one-hybrid and two-hybrid methods. We tested our system by selecting zinc finger variants (from a large randomized library) that bind tightly and specifically to desired DNA target sites. Our method allows sequence-specific zinc fingers to be isolated in a single selection step, and thus it should be more rapid than phage display strategies that typically require multiple enrichment/amplification cycles. Given the large library sizes our bacterial-based selection system can handle, this method should provide a powerful tool for identifying and optimizing protein-DNA and protein-protein interactions.
Figures

Similar articles
-
Identifying and modifying protein-DNA and protein-protein interactions using a bacterial two-hybrid selection system.J Cell Biochem Suppl. 2001;Suppl 37:53-7. doi: 10.1002/jcb.10065. J Cell Biochem Suppl. 2001. PMID: 11842428 Review.
-
Engineering Cys2His2 zinc finger domains using a bacterial cell-based two-hybrid selection system.Methods Mol Biol. 2007;408:317-34. doi: 10.1007/978-1-59745-547-3_17. Methods Mol Biol. 2007. PMID: 18314590
-
Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers.Structure. 2000 Jul 15;8(7):739-50. doi: 10.1016/s0969-2126(00)00161-1. Structure. 2000. PMID: 10903945
-
Engineering single Cys2His2 zinc finger domains using a bacterial cell-based two-hybrid selection system.Methods Mol Biol. 2010;649:31-50. doi: 10.1007/978-1-60761-753-2_2. Methods Mol Biol. 2010. PMID: 20680826
-
One from column A and two from column B: the benefits of phage display in molecular-recognition studies.Curr Opin Chem Biol. 2002 Feb;6(1):92-6. doi: 10.1016/s1367-5931(01)00287-3. Curr Opin Chem Biol. 2002. PMID: 11827830 Review.
Cited by
-
Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment.PLoS Comput Biol. 2015 Feb 3;11(2):e1004067. doi: 10.1371/journal.pcbi.1004067. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25646976 Free PMC article.
-
Prediction of DNA-binding specificity in zinc finger proteins.J Biosci. 2012 Jul;37(3):483-91. doi: 10.1007/s12038-012-9213-7. J Biosci. 2012. PMID: 22750985
-
Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases.Nucleic Acids Res. 2012 Apr;40(8):3741-52. doi: 10.1093/nar/gkr1214. Epub 2011 Dec 14. Nucleic Acids Res. 2012. PMID: 22169954 Free PMC article.
-
Programmable CRISPR-Cas transcriptional activation in bacteria.Mol Syst Biol. 2020 Jul;16(7):e9427. doi: 10.15252/msb.20199427. Mol Syst Biol. 2020. PMID: 32657546 Free PMC article.
-
Zinc finger nucleases as tools to understand and treat human diseases.BMC Med. 2010 Jul 1;8:42. doi: 10.1186/1741-7015-8-42. BMC Med. 2010. PMID: 20594338 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

