A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID
- PMID: 10852950
- PMCID: PMC16519
- DOI: 10.1073/pnas.120074297
A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID
Abstract
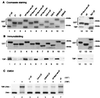
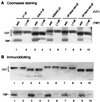
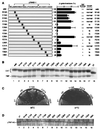
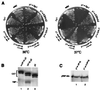
The TATA box-binding activity of transcription factor IID (TFIID) is autoinhibited by the N-terminal domain of the Drosophila TATA box-binding protein- (TBP) associated factor 230/yeast TBP-associated factor 145 subunit, which binds to the TATA box-binding domain of TBP by mimicking the TATA box structure. Here, we propose a mechanism of transcriptional activation that involves antirepression of this autoinhibitory activity by transcriptional activators. Like the autoinhibitory domain of TFIID, various acidic activators interact with the TATA box-binding domain of TBP. Moreover, the autoinhibitory domain of TFIID, which is known to interact with only the TATA box-binding domain of TBP, acts as an activation domain when fused to the GAL4 DNA-binding domain, indicating that interaction with the TATA-binding domain of TBP is crucial for activation of transcription. In a reciprocal fashion, the acidic activation domains can function as the autoinhibitory domain when the latter is replaced by the former within TFIID. These results indicate that activation domains and the autoinhibitory domain of TFIID are interchangeable, supporting a role for transcriptional activators as antirepressors of the autoinhibitory activity of the TATA box binding of TFIID.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

