An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana
- PMID: 10854410
- PMCID: PMC310908
- DOI: 10.1101/gr.10.6.776
An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana
Abstract
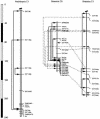
A detailed comparative map of Brassica oleracea and Arabidopsis thaliana has been established based largely on mapping of Arabidopsis ESTs in two Arabidopsis and four Brassica populations. Based on conservative criteria for inferring synteny, "one to one correspondence" between Brassica and Arabidopsis chromosomes accounted for 57% of comparative loci. Based on 186 corresponding loci detected in B. oleracea and A. thaliana, at least 19 chromosome structural rearrangements differentiate B. oleracea and A. thaliana orthologs. Chromosomal duplication in the B. oleracea genome was strongly suggested by parallel arrangements of duplicated loci on different chromosomes, which accounted for 41% of loci mapped in Brassica. Based on 367 loci mapped, at least 22 chromosomal rearrangements differentiate B. oleracea homologs from one another. Triplication of some Brassica chromatin and duplication of some Arabidopsis chromatin were suggested by data that could not be accounted for by the one-to-one and duplication models, respectively. Twenty-seven probes detected three or more loci in Brassica, which represent 25.3% of the 367 loci mapped in Brassica. Thirty-one probes detected two or more loci in Arabidopsis, which represent 23.7% of the 262 loci mapped in Arabidopsis. Application of an EST-based, cross-species genomic framework to isolation of alleles conferring phenotypes unique to Brassica, as well as the challenges and opportunities in extrapolating genetic information from Arabidopsis to Brassica and to more distantly related crops, are discussed.
Figures





References
-
- Agyare FD, Lashkari DA, Lagos A, Namath AF, Lagos G, et al. Mapping expressed sequence tag sites on yeast artificial chromosome clones of Arabidopsis thaliana DNA. Genome Res. 1997;7:1–9. - PubMed
-
- Arumuganathan K, Earle ED. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep. 1991;9:208–218.
-
- Berry R, Stevens TJ, Walter NAR, Wilcox AS, Rubano T, et al. Gene-based sequence-tagged-sites (STSs) as the basis for a human gene map. Nat Genet. 1995;10:415–423. - PubMed
-
- Botella MA, Coleman MJ, Hughes DE, Nishimura MT, Jones JDG, et al. Map positions of 47 Arabidopsis sequences with sequence similarity to disease resistance genes. Plant J. 1997;12:1197–1211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
