The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome
- PMID: 10854413
- PMCID: PMC310893
- DOI: 10.1101/gr.10.6.819
The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome
Abstract
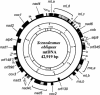
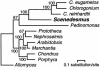
Two distinct mitochondrial genome types have been described among the green algal lineages investigated to date: a reduced-derived, Chlamydomonas-like type and an ancestral, Prototheca-like type. To determine if this unexpected dichotomy is real or is due to insufficient or biased sampling and to define trends in the evolution of the green algal mitochondrial genome, we sequenced and analyzed the mitochondrial DNA (mtDNA) of Scenedesmus obliquus. This genome is 42,919 bp in size and encodes 42 conserved genes (i.e., large and small subunit rRNA genes, 27 tRNA and 13 respiratory protein-coding genes), four additional free-standing open reading frames with no known homologs, and an intronic reading frame with endonuclease/maturase similarity. No 5S rRNA or ribosomal protein-coding genes have been identified in Scenedesmus mtDNA. The standard protein-coding genes feature a deviant genetic code characterized by the use of UAG (normally a stop codon) to specify leucine, and the unprecedented use of UCA (normally a serine codon) as a signal for termination of translation. The mitochondrial genome of Scenedesmus combines features of both green algal mitochondrial genome types: the presence of a more complex set of protein-coding and tRNA genes is shared with the ancestral type, whereas the lack of 5S rRNA and ribosomal protein-coding genes as well as the presence of fragmented and scrambled rRNA genes are shared with the reduced-derived type of mitochondrial genome organization. Furthermore, the gene content and the fragmentation pattern of the rRNA genes suggest that this genome represents an intermediate stage in the evolutionary process of mitochondrial genome streamlining in green algae.
Figures


Similar articles
-
DNA sequence analysis of the complete mitochondrial genome of the green alga Scenedesmus obliquus: evidence for UAG being a leucine and UCA being a non-sense codon.Gene. 2000 Jul 25;253(1):13-8. doi: 10.1016/s0378-1119(00)00228-6. Gene. 2000. PMID: 10925198
-
The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae.Mol Biol Evol. 2004 May;21(5):922-35. doi: 10.1093/molbev/msh099. Epub 2004 Mar 10. Mol Biol Evol. 2004. PMID: 15014170
-
The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants.Mol Biol Evol. 2002 Jan;19(1):24-38. doi: 10.1093/oxfordjournals.molbev.a003979. Mol Biol Evol. 2002. PMID: 11752187
-
Evolution of the mitochondrial genome: protist connections to animals, fungi and plants.Curr Opin Microbiol. 2004 Oct;7(5):528-34. doi: 10.1016/j.mib.2004.08.008. Curr Opin Microbiol. 2004. PMID: 15451509 Review.
-
The Drosophila mitochondrial genome.Oxf Surv Eukaryot Genes. 1984;1:1-35. Oxf Surv Eukaryot Genes. 1984. PMID: 6400770 Review.
Cited by
-
PLMItRNA, a database for mitochondrial tRNA genes and tRNAs in photosynthetic eukaryotes.Nucleic Acids Res. 2001 Jan 1;29(1):167-8. doi: 10.1093/nar/29.1.167. Nucleic Acids Res. 2001. PMID: 11125079 Free PMC article.
-
The complete mitochondrial DNA sequence of the green algae Hariotina sp. F30 (Scenedesmaceae, Sphaeropleales, Chlorophyceae).Mitochondrial DNA B Resour. 2016 Mar 28;1(1):124-125. doi: 10.1080/23802359.2016.1144089. Mitochondrial DNA B Resour. 2016. PMID: 33473431 Free PMC article.
-
A single mutation in the first transmembrane domain of yeast COX2 enables its allotopic expression.Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):5047-52. doi: 10.1073/pnas.1000735107. Epub 2010 Mar 1. Proc Natl Acad Sci U S A. 2010. PMID: 20194738 Free PMC article.
-
Mitochondrial genome sequence evolution in Chlamydomonas.Genetics. 2007 Feb;175(2):819-26. doi: 10.1534/genetics.106.063156. Epub 2006 Dec 6. Genetics. 2007. PMID: 17151253 Free PMC article.
-
Genome sequencing, assembly, and annotation of the self-flocculating microalga Scenedesmus obliquus AS-6-11.BMC Genomics. 2020 Oct 27;21(1):743. doi: 10.1186/s12864-020-07142-4. BMC Genomics. 2020. PMID: 33109102 Free PMC article.
References
-
- Adachi J, Hasegawa M. Computer science monographs, No. 28. MOLPHY version 2.3B3: Programs for molecular phylogenetics based on maximum likelihood. Tokyo: Institute of Statistical Mathematics; 1996.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Boer PH, Gray MW. Transfer RNA genes and the genetic code in Chlamydomonas reinhardtii mitochondria. Curr Genet. 1988a;14:583–590. - PubMed
-
- Boer PH, Gray MW. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988b;55:399–411. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
