Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion
- PMID: 10856303
- PMCID: PMC2552403
- DOI: 10.1074/jbc.M004008200
Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion
Abstract
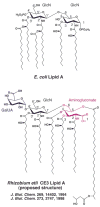
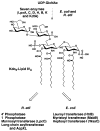
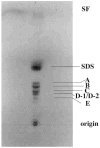
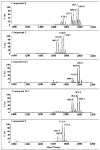
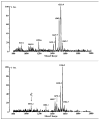
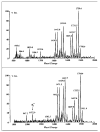
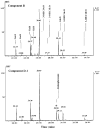
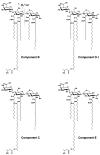
Lipid A of Rhizobium etli CE3 differs dramatically from that of other Gram-negative bacteria. Key features include the presence of an unusual C28 acyl chain, a galacturonic acid moiety at position 4', and an acylated aminogluconate unit in place of the proximal glucosamine. In addition, R. etli lipid A is reported to lack phosphate and acyloxyacyl residues. Most of these remarkable structural claims are consistent with our recent enzymatic studies. However, the proposed R. etli lipid A structure is inconsistent with the ability of the precursor (3-deoxy-D-manno-octulosonic acid)(2)-4'-(32)P-lipid IV(A) to accept a C28 chain in vitro (Brozek, K. A., Carlson, R. W., and Raetz, C. R. H. (1996) J. Biol. Chem. 271, 32126-32136). To re-evaluate the structure, CE3 lipid A was isolated by new chromatographic procedures. CE3 lipid A is now resolved into six related components. Aminogluconate is present in D-1, D-2, and E, whereas B and C contain the typical glucosamine disaccharide seen in lipid A of most other bacteria. All the components possess a peculiar acyloxyacyl moiety at position 2', which includes the ester-linked C28 chain. As judged by mass spectrometry, the distal glucosamine units of A through E are the same, but the proximal units are variable. As described in the accompanying article (Que, N. L. S., Ribeiro, A. A., and Raetz, C. R. H. (2000) J. Biol. Chem. 275, 28017-28027), the discovery of component B suggests a plausible enzymatic pathway for the biosynthesis of the aminogluconate residue found in species D-1, D-2, and E of R. etli lipid A. We suggest that the unusual lipid A species of R. etli might be essential during symbiosis with leguminous host plants.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

