Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15
- PMID: 10858228
- PMCID: PMC101706
- DOI: 10.1128/IAI.68.7.4108-4116.2000
Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15
Abstract
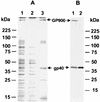
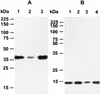
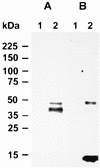
Cryptosporidium parvum is a significant cause of diarrheal disease worldwide. The specific molecules that mediate C. parvum-host cell interactions and the molecular mechanisms involved in the pathogenesis of cryptosporidiosis are unknown. In this study we have shown that gp40, a mucin-like glycoprotein, is localized to the surface and apical region of invasive stages of the parasite and is shed from its surface. gp40-specific antibodies neutralize infection in vitro, and native gp40 binds specifically to host cells, implicating this glycoprotein in C. parvum attachment to and invasion of host cells. We have cloned and sequenced a gene designated Cpgp40/15 that encodes gp40 as well as gp15, an antigenically distinct, surface glycoprotein also implicated in C. parvum-host cell interactions. Analysis of the deduced amino acid sequence of the 981-bp Cpgp40/15 revealed the presence of an N-terminal signal peptide, a polyserine domain, multiple predicted O-glycosylation sites, a single potential N-glycosylation site, and a hydrophobic region at the C terminus, a finding consistent with what is required for the addition of a GPI anchor. There is a single copy of Cpgp40/15 in the C. parvum genome, and this gene does not contain introns. Our data indicate that the two Cpgp40/15-encoded proteins, gp40 and gp15, are products of proteolytic cleavage of a 49-kDa precursor protein which is expressed in intracellular stages of the parasite. The surface localization of gp40 and gp15 and their involvement in the host-parasite interaction suggest that either or both of these glycoproteins may serve as effective targets for specific preventive or therapeutic measures for cryptosporidiosis.
Figures




References
-
- Barnes D A, Bonnin A, Huang J X, Gousset L, Wu J, Gut J, Doyle P, Dubremetz J F, Ward H, Petersen C. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol. 1998;96:93–110. - PubMed
-
- Blackman M, Holder A. Secondary processing of the Plasmodium falciparum merozoite surface protein (MSP-1) by a calcium-dependent membrane bound serine protease: shedding of MSP-133 as a non-covalently associated complex with other fragments of MSP-1. Mol Biochem Parasitol. 1992;50:307–316. - PubMed
-
- Bradley P J, Boothroyd J C. Identification of the pro-mature processing site of Toxoplasma ROP1 by mass spectrometry. Mol Biochem Parasitol. 1999;100:103–109. - PubMed
-
- Eng J, McCormack A L, Yates J R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

