Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families
- PMID: 10858232
- PMCID: PMC101716
- DOI: 10.1128/IAI.68.7.4155-4168.2000
Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families
Abstract
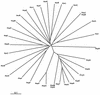
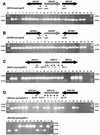
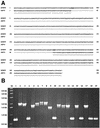
The two complete genomic sequences of Helicobacter pylori J99 and 26695 were used to compare the paralogous families (related genes within one genome, likely to have related function) of genes predicted to encode outer membrane proteins which were present in each strain. We identified five paralogous gene families ranging in size from 3 to 33 members; two of these families contained members specific for either H. pylori J99 or H. pylori 26695. Most orthologous protein pairs (equivalent genes between two genomes, same function) shared considerable identity between the two strains. The unusual set of outer membrane proteins and the specialized outer membrane may be a reflection of the adaptation of H. pylori to the unique gastric environment where it is found. One subfamily of proteins, which contains both channel-forming and adhesin molecules, is extremely highly related at the sequence level and has likely arisen due to ancestral gene duplication. In addition, the largest paralogous family contained two essentially identical pairs of genes in both strains. The presence and genomic organization of these two pairs of duplicated genes were analyzed in a panel of independent H. pylori isolates. While one pair was present in every strain examined, one allele of the other pair appeared partially deleted in several isolates.
Figures



References
-
- Akopyants N S, Jiang Q, Taylor D E, Berg D E. Corrected identity of isolates of Helicobacter pylori reference strain NCTC11637. Helicobacter. 1997;2:48–52. - PubMed
-
- Alm R A, Ling L L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:186–190. - PubMed
-
- Appelmelk B J, Martin S L, Monteiro M A, Clayton C A, McColm A A, Zheng P, Verboom T, Maaskant J J, van den Eijnden D H, Hokke C H, Perry M B, Vandenbroucke-Grauls C M J E, Kusters J G. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect Immun. 1999;67:5361–5366. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

