Correlation between pretreatment levels of interferon response genes and clinical responses to an immune response modifier (Imiquimod) in genital warts
- PMID: 10858346
- PMCID: PMC89977
- DOI: 10.1128/AAC.44.7.1869-1873.2000
Correlation between pretreatment levels of interferon response genes and clinical responses to an immune response modifier (Imiquimod) in genital warts
Abstract
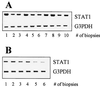
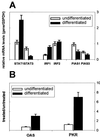
Imiquimod (IQ) has been successfully used in treatment of genital warts. In clinical settings, patients responded well but wart reduction rates varied. Our aim was to find a correlation between clinical responses and pretreatment (constitutive) levels of genes that might be involved in the molecular action of IQ. Since IQ is a cytokine inducer, we analyzed levels of expression of genes of the JAK/STAT signaling pathway and their inhibitors as well as interferon response factors (IRFs) in pretreatment biopsy specimens from complete responders (99 to 100% wart reduction rate) versus incomplete responders (75 to 92% wart reduction rate) by reverse transcription-PCR. We found that mRNA levels of signal transducer and activator of transcription 1 (STAT1) and IRF1 were higher in complete responders than in incomplete responders. Incomplete responders expressed larger amounts of STAT3, IRF2, and protein inhibitor of activated STAT1 (PIAS1) mRNAs compared to complete responders before IQ treatment. We hypothesize that high-level expression of STAT1 and IRF1 is advantageous for a better IQ response. The observed differences in constitutive mRNA levels of these genes may be the consequence of alterations in cellular differentiation and/or variable expression of endogenous interferons. Previous in vitro studies showed that keratinocyte differentiation coordinates the balance between positive and negative signals along the JAK/STAT pathway by regulating the IRF1:IRF2 and STAT1:PIAS1 ratios and thus affecting induction of IQ-inducible genes. Specifically, differentiation supports constitutive expression of STAT1 and IRF1 mRNAs but not expression of IRF2 and PIAS1. Our data are in good agreement with studies that showed the importance of STAT1 in cytokine induction and activation of interferon-responsive genes by IQ.
Figures



References
-
- Anonymous. Imiquimod for genital warts. Med Lett Drugs Ther. 1997;39:118–119. - PubMed
-
- Arany I, Brysk M M, Brysk H, Tyring S K. Response to interferon treatment decreases with epidermal dedifferentiation in condylomas. Antivir Res. 1996;32:19–26. - PubMed
-
- Arany I, Tyring S K. Activation of local cell-mediated immunity in interferon responsive patients with human papillomavirus-associated lesions. J Interferon Cytokine Res. 1996;16:453–460. - PubMed
-
- Arany I, Tyring S K. Status of local cellular immunity in interferon responsive and nonresponsive human papillomavirus-associated lesions. Sex Transm Dis. 1996;26:475–480. - PubMed
-
- Beutner K R, Spruance S L, Hougham A J, Fox T L, Owens M L, Douglas J M., Jr Treatment of genital warts with an immune-response modifier (imiquimod) J Am Acad Dermatol. 1998;38:230–239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

