Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum
- PMID: 10858348
- PMCID: PMC89979
- DOI: 10.1128/AAC.44.7.1878-1886.2000
Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum
Abstract
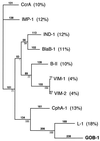
Although the carbapenem-hydrolyzing beta-lactamase (CHbetaL) BlaB-1 is known to be in Chryseobacterium meningosepticum NCTC 10585, a second CHbetaL gene, bla(GOB-1), was cloned from another C. meningosepticum clinical isolate (PINT). The G+C content of bla(GOB-1) (36%) indicated the likely chromosomal origin of this gene. Its expression in Escherichia coli DH10B yields a mature CHbetaL with a pI of 8.7 and a relative molecular mass of 28.2 kDa. In E. coli, GOB-1 conferred resistance to narrow-spectrum cephalosporins and reduced susceptibility to ureidopenicillins, broad-spectrum cephalosporins, and carbapenems. GOB-1 had a broad-spectrum hydrolysis profile including penicillins and cephalosporins (but not aztreonam). The catalytic efficiency for meropenem was higher than for imipenem. GOB-1 had low amino acid identity with the class B CHbetaLs, sharing 18% with the closest, L-1 from Stenotrophomonas maltophilia, and only 11% with BlaB-1. Most of the conserved amino acids that may be involved in the active site of CHbetaLs (His-101, Asp-103, His-162, and His-225) were identified in GOB-1. Sequence heterogeneity was found for GOB-1-like and BlaB-1-like beta-lactamases, having 90 to 100% and 86 to 100% amino acid identity, respectively, among 10 unrelated C. meningosepticum isolates. Each isolate had a GOB-1-like and a BlaB-1-like gene. The same combination of GOB-1-like and BlaB-1-like beta-lactamases was not found in two different isolates. C. meningosepticum is a bacterial species with two types of unrelated chromosome-borne class B CHbetaLs that can be expressed in E. coli and, thus, may represent a clinical threat if spread in gram-negative aerobes.
Figures




References
-
- Ambler R P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. - PubMed
-
- Bellais S, Léotard S, Poirel L, Naas T, Nordmann P. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett. 1999;171:127–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

