A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil
- PMID: 10858358
- PMCID: PMC89989
- DOI: 10.1128/AAC.44.7.1936-1942.2000
A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil
Abstract
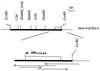
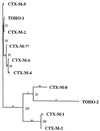
To estimate the diversity of extended-spectrum beta-lactamases in Brazil, 18 strains from different species of the family Enterobacteriaceae exhibiting a positive double-disk synergy test were collected by a clinical laboratory from several hospitals in Rio de Janeiro, Brazil, in 1996 and 1997. Four strains (Proteus mirabilis, Enterobacter cloacae, Enterobacter aerogenes, and Citrobacter amalonaticus) hybridized with a 550-bp CTX-M probe. The P. mirabilis strain produced a CTX-M-2 enzyme. The E. cloacae, E. aerogenes, and C. amalonaticus isolates harbored a bla gene which was identified by cloning and sequencing as a bla(CTX-M) gene. E. coli HB101 transconjugants and the E. coli DH5alpha transformant harboring a recombinant plasmid produced a CTX-M beta-lactamase with an isoelectric point of 7.6 conferring a resistance phenotype characterized by a higher level of resistance to cefotaxime than to ceftazidime, as observed with the other CTX-M enzymes. The deduced protein sequence showed a novel Ambler class A CTX-M enzyme, named CTX-M-8, which had 83 to 88% identity with the previously described CTX-M enzymes. The phylogenic study of the CTX-M family including CTX-M-8 revealed four CTX-M types, CTX-M-8 being the first member of a new phylum of CTX-M enzymes. The evolutionary distances between the four types of CTX-M were large, suggesting that the four clusters branched off early from a distant unknown enzyme and that intermediate enzymes probably existed.
Figures


References
-
- Ballow C, Schentag J J. Trends in antibiotic utilization and bacterial resistance. Report of the National Nosocomial Resistance Surveillance Group. Diagn Microbiol Infect Dis. 1992;15:37S–42S. - PubMed
-
- Barthelemy M, Peduzzi J, Bernard H, Tancrede C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. - PubMed
-
- Bauernfeind A, Casellas J M, Goldberg M, Holley M, Jungwirth R, Mangold P, Rohnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

