Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading
- PMID: 10859170
- PMCID: PMC316685
Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading
Abstract
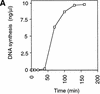
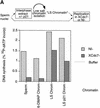
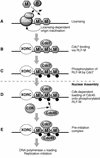
The assembly and disassembly of protein complexes at replication origins play a crucial role in the regulation of chromosomal DNA replication. The sequential binding of the origin recognition complex (ORC), Cdc6, and the minichromosome maintenance (MCM/P1) proteins produces a licensed replication origin. Before the initiation of replication can occur, each licensed origin must be acted upon by S phase-inducing CDKs and the Cdc7 protein kinase. In the present report we describe the role of Xenopus Cdc7 (XCdc7) in DNA replication using cell-free extracts of Xenopus eggs. We show that XCdc7 binds to chromatin during G(1) and S phase. XCdc7 associates with chromatin only once origins have been licensed, but this association does not require the continued presence of XORC or XCdc6 once they have fulfilled their essential role in licensing. Moreover, XCdc7 is required for the subsequent CDK-dependent loading of XCdc45 but is not required for the destabilization of origins that occurs once licensing is complete. Finally, we show that CDK activity is not necessary for XCdc7 to associate with chromatin, induce MCM/P1 phosphorylation, or perform its essential replicative function. From these results we suggest a simple model for the assembly of functional initiation complexes in the Xenopus system.
Figures












Similar articles
-
Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk.EMBO J. 1998 Oct 1;17(19):5699-707. doi: 10.1093/emboj/17.19.5699. EMBO J. 1998. PMID: 9755170 Free PMC article.
-
Analysis of Cdc6 function in the assembly of mammalian prereplication complexes.Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1347-52. doi: 10.1073/pnas.032677499. Epub 2002 Jan 22. Proc Natl Acad Sci U S A. 2002. PMID: 11805305 Free PMC article.
-
A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication.BMC Mol Biol. 2004 Jun 28;5:5. doi: 10.1186/1471-2199-5-5. BMC Mol Biol. 2004. PMID: 15222894 Free PMC article.
-
Cdc7 kinase complex: a key regulator in the initiation of DNA replication.J Cell Physiol. 2002 Mar;190(3):287-96. doi: 10.1002/jcp.10070. J Cell Physiol. 2002. PMID: 11857444 Review.
-
Control of DNA replication licensing in a cell cycle.Genes Cells. 2002 Jun;7(6):523-34. doi: 10.1046/j.1365-2443.2002.00544.x. Genes Cells. 2002. PMID: 12059957 Review.
Cited by
-
Cdc45 is a critical effector of myc-dependent DNA replication stress.Cell Rep. 2013 May 30;3(5):1629-39. doi: 10.1016/j.celrep.2013.04.002. Epub 2013 May 2. Cell Rep. 2013. PMID: 23643534 Free PMC article.
-
Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication.J Cell Biol. 2001 Mar 19;152(6):1267-78. doi: 10.1083/jcb.152.6.1267. J Cell Biol. 2001. PMID: 11257126 Free PMC article.
-
The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding.EMBO J. 2005 Dec 21;24(24):4334-44. doi: 10.1038/sj.emboj.7600892. Epub 2005 Nov 24. EMBO J. 2005. PMID: 16369567 Free PMC article.
-
Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin.EMBO J. 2002 Mar 15;21(6):1437-46. doi: 10.1093/emboj/21.6.1437. EMBO J. 2002. PMID: 11889049 Free PMC article.
-
DDK: The Outsourced Kinase of Chromosome Maintenance.Biology (Basel). 2022 Jun 7;11(6):877. doi: 10.3390/biology11060877. Biology (Basel). 2022. PMID: 35741398 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
