PapD-like chaperones provide the missing information for folding of pilin proteins
- PMID: 10859353
- PMCID: PMC16609
- DOI: 10.1073/pnas.130183897
PapD-like chaperones provide the missing information for folding of pilin proteins
Abstract
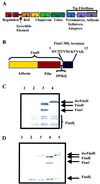
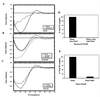
A fundamental question in molecular biology is how proteins fold into domains that can serve as assembly modules for building up large macromolecular structures. The biogenesis of pili on the surface of Gram-negative bacteria requires the orchestration of a complex process that includes protein synthesis, folding via small chaperones, secretion, and assembly. The results presented here support the hypothesis that pilus subunit folding and biogenesis proceed via mechanisms termed donor strand complementation and donor strand exchange. Here we show that the steric information necessary for pilus subunit folding is not contained in one polypeptide sequence. Rather, the missing information is transiently donated by a strand of a small chaperone to allow folding. Providing the missing information for folding, via a 13-amino acid peptide extension to the C-terminal end of a pilus subunit, resulted in the production of a protein that no longer required the chaperone to fold. This mechanism of small periplasmic chaperone function described here deviates from classical hsp60 chaperone-assisted folding.
Figures


Comment in
-
Anfinsen comes out of the cage during assembly of the bacterial pilus.Proc Natl Acad Sci U S A. 2000 Jul 5;97(14):7670-2. doi: 10.1073/pnas.97.14.7670. Proc Natl Acad Sci U S A. 2000. PMID: 10884397 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

