Maize as a model for the evolution of plant nuclear genomes
- PMID: 10860964
- PMCID: PMC34377
- DOI: 10.1073/pnas.97.13.7008
Maize as a model for the evolution of plant nuclear genomes
Abstract
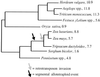
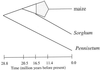
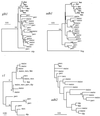
The maize genome is replete with chromosomal duplications and repetitive DNA. The duplications resulted from an ancient polyploid event that occurred over 11 million years ago. Based on DNA sequence data, the polyploid event occurred after the divergence between sorghum and maize, and hence the polyploid event explains some of the difference in DNA content between these two species. Genomic rearrangement and diploidization followed the polyploid event. Most of the repetitive DNA in the maize genome is retrotransposable elements, and they comprise 50% of the genome. Retrotransposon multiplication has been relatively recent-within the last 5-6 million years-suggesting that the proliferation of retrotransposons has also contributed to differences in DNA content between sorghum and maize. There are still unanswered questions about repetitive DNA, including the distribution of repetitive DNA throughout the genome, the relative impacts of retrotransposons and chromosomal duplication in plant genome evolution, and the hypothesized correlation of duplication events with transposition. Population genetic processes also affect the evolution of genomes. We discuss how centromeric genes should, in theory, contain less genetic diversity than noncentromeric genes. In addition, studies of diversity in the wild relatives of maize indicate that different genes have different histories and also show that domestication and intensive breeding have had heterogeneous effects on genetic diversity across genes.
Figures

References
-
- Stebbins G L. Variation and Evolution in Plants. New York: Columbia Univ. Press; 1950.
-
- Bennett M D, Leitch I J. Ann Bot. 1995;76:113–176.
-
- McClintock B. Z Zellforsch Mikrosk Anat. 1933;19:191–237.
-
- Ting Y C. Cytologia. 1966;31:324–329.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

