An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus
- PMID: 10864633
- PMCID: PMC112129
- DOI: 10.1128/jvi.74.14.6242-6250.2000
An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus
Abstract
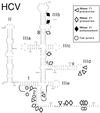
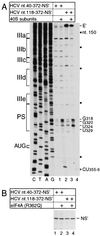
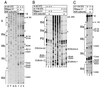
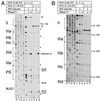
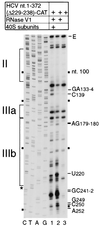
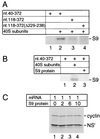
Hepatitis C virus translation is initiated on a approximately 330-nucleotide (nt)-long internal ribosomal entry site (IRES) at the 5' end of the genome. In this process, a 43S preinitiation complex (comprising a 40S ribosomal subunit, eukaryotic initiation factor 3 (eIF3), and a ternary [eIF2-GTP-initiator tRNA] complex) binds the IRES in a precise manner so that the initiation codon is placed at the ribosomal P site. This binding step involves specific interactions between the IRES and different components of the 43S complex. The 40S subunit and eIF3 can bind to the IRES independently; previous analyses revealed that eIF3 binds specifically to an apical half of IRES domain III. Nucleotides in the IRES that are involved in the interaction with the 40S subunit were identified by RNase footprinting and mapped to the basal half of domain III and in domain IV. Interaction sites were identified in locations that have been found to be essential for IRES function, including (i) the apical loop residues GGG(266-268) in subdomain IIId and (ii) the pseudoknot. Extensive protection from RNase cleavage also occurred downstream of the pseudoknot in domain IV, flanking both sides of the initiation codon and corresponding in length to that of the mRNA-binding cleft of the 40S subunit. These results indicate that the 40S subunit makes multiple interactions with the IRES and suggest that only nucleotides in domain IV are inserted into the mRNA-binding cleft of the 40S subunit.
Figures
References
-
- Becher P, Orlich M, Shannon A D, Horner G, König M, Thiel J-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78:1357–1366. - PubMed
-
- Browning K S, Leung D W, Clark J M. Protection of satellite tobacco necrosis virus ribonucleic acid by wheat germ 40S and 80S ribosomes. Biochemistry. 1980;19:2276–2283. - PubMed
-
- Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

