Postcleavage sequence specificity in V(D)J recombination
- PMID: 10866660
- PMCID: PMC85953
- DOI: 10.1128/MCB.20.14.5032-5040.2000
Postcleavage sequence specificity in V(D)J recombination
Abstract
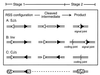
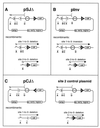
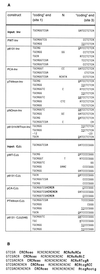
Unintended DNA rearrangements in a differentiating lymphocyte can have severe, oncogenic consequences, but the mechanisms for avoiding pathogenic outcomes in V(D)J recombination are not well understood. The first level at which fidelity is instituted is in discrimination by the recombination proteins between authentic and inauthentic recombination signal sequences. Nevertheless, this discrimination is not absolute and cannot fully eliminate targeting errors. To learn more about the basis of specificity during V(D)J recombination, we have investigated whether it is possible for the recombination machinery to detect an inaccurately targeted sequence subsequent to cleavage. These studies indicate that even postcleavage steps in V(D)J recombination are sequence specific and that noncanonical sequences will not efficiently support the resolution of recombination intermediates in vivo. Accordingly, interventions after a mistargeting event conceivably occur at a late stage in the joining process and the likelihood may well be crucial to enforcing fidelity during antigen receptor gene rearrangement.
Figures


References
-
- Agrawal A, Schatz D B. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. - PubMed
-
- Aplan P D, Lombardi D P, Ginsberg A M, Cossman J, Bertness V L, Kirsch I R. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. - PubMed
-
- Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by the recombination activating genes RAG1 and RAG2. Mol Cell. 1998;2:817–828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
