A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein
- PMID: 10866662
- PMCID: PMC85955
- DOI: 10.1128/MCB.20.14.5048-5063.2000
A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein
Abstract
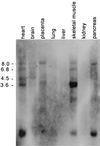
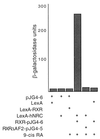
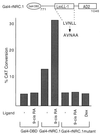
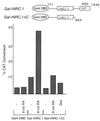
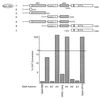
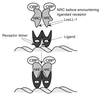
We describe the cloning and characterization of a new family of nuclear receptor coregulators (NRCs) which modulate the function of nuclear hormone receptors in a ligand-dependent manner. NRCs are expressed as alternatively spliced isoforms which may exhibit different intrinsic activities and receptor specificities. The NRCs are organized into several modular structures and contain a single functional LXXLL motif which associates with members of the steroid hormone and thyroid hormone/retinoid receptor subfamilies with high affinity. Human NRC (hNRC) harbors a potent N-terminal activation domain (AD1), which is as active as the herpesvirus VP16 activation domain, and a second activation domain (AD2) which overlaps with the receptor-interacting LXXLL region. The C-terminal region of hNRC appears to function as an inhibitory domain which influences the overall transcriptional activity of the protein. Our results suggest that NRC binds to liganded receptors as a dimer and this association leads to a structural change in NRC resulting in activation. hNRC binds CREB-binding protein (CBP) with high affinity in vivo, suggesting that hNRC may be an important functional component of a CBP complex involved in mediating the transcriptional effects of nuclear hormone receptors.
Figures







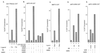

References
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Carson-Jurica M A, Schrader W T, O'Malley B W. Steroid receptor family: structure and functions. Endocrine Rev. 1990;11:201–218. - PubMed
-
- Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
