A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113
- PMID: 10869066
- PMCID: PMC94573
- DOI: 10.1128/JB.182.14.3913-3919.2000
A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113
Abstract
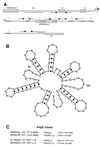
The GacS-GacA two-component signal transduction system, which is highly conserved in gram-negative bacteria, is required for the production of exoenzymes and secondary metabolites in Pseudomonas spp. Screening of a Pseudomonas fluorescens F113 gene bank led to the isolation of a previously undefined locus which could restore secondary metabolite production to both gacS and gacA mutants of F113. Sequence analysis of this locus demonstrated that it did not contain any obvious Pseudomonas protein-coding open reading frames or homologues within available databases. Northern analysis indicated that the locus encodes an RNA (PrrB RNA) which is able to phenotypically complement gacS and gacA mutants and is itself regulated by the GacS-GacA two-component signal transduction system. Primer extension analysis of the 132-base transcript identified the transcription start site located downstream of a sigma(70) promoter sequence from positions -10 to -35. Inactivation of the prrB gene in F113 resulted in a significant reduction of 2, 4-diacetylphloroglucinol (Phl) and hydrogen cyanide (HCN) production, while increased metabolite production was observed when prrB was overexpressed. The prrB gene sequence contains a number of imperfect repeats of the consensus sequence 5'-AGGA-3', and sequence analysis predicted a complex secondary structure featuring multiple putative stem-loops with the consensus sequences predominantly positioned at the single-stranded regions at the ends of the stem-loops. This structure is similar to the CsrB and RsmB regulatory RNAs in Escherichia coli and Erwinia carotovora, respectively. Results suggest that a regulatory RNA molecule is involved in GacA-GacS-mediated regulation of Phl and HCN production in P. fluorescens F113.
Figures



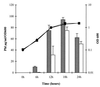
 ) and F113prrB (▭) is shown by the bars and the left y axis, and OD600 of wild-type F113 (■) and F113prrB (⧫) is shown by the curves and the right y axis.
) and F113prrB (▭) is shown by the bars and the left y axis, and OD600 of wild-type F113 (■) and F113prrB (⧫) is shown by the curves and the right y axis.References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic local alignment tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Chatterjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

