Integration and excision of a Bacteroides conjugative transposon, CTnDOT
- PMID: 10869083
- PMCID: PMC94590
- DOI: 10.1128/JB.182.14.4035-4043.2000
Integration and excision of a Bacteroides conjugative transposon, CTnDOT
Abstract
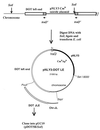
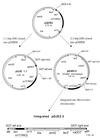
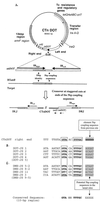
Bacteroides conjugative transposons (CTns) are thought to transfer by first excising themselves from the chromosome to form a nonreplicating circle, which is then transferred by conjugation to a recipient. Earlier studies showed that transfer of most Bacteroides CTns is stimulated by tetracycline, but it was not known which step in transfer is regulated. We have cloned and sequenced both ends of the Bacteroides CTn, CTnDOT, and have used this information to examine excision and integration events. A segment of DNA that contains the joined ends of CTnDOT and an adjacent open reading frame (ORF), intDOT, was necessary and sufficient for integration into the Bacteroides chromosome. Integration of this miniature form of the CTn was not regulated by tetracycline. Excision of CTnDOT and formation of the circular intermediate were detected by PCR, using primers designed from the end sequences. Sequence analysis of the PCR products revealed that excision and integration involve a 5-bp coupling sequence-type mechanism possibly similar to that used by CTn Tn916, a CTn found originally in enterococci. PCR analysis also demonstrated that excision is a tetracycline-regulated step in transfer. The integrated minielement containing intDOT and the ends of CTnDOT did not excise, nor did a larger minielement that also contained an ORF located immediately downstream of intDOT designated orf2. Thus, excision involves other genes besides intDOT and orf2. Both intDOT and orf2 were disrupted by single-crossover insertions. Analysis of the disruption mutants showed that intDOT was essential for excision but orf2 was not. Despite its proximity to the integrase gene, orf2 appears not to be essential for excision.
Figures


References
-
- Caparon M G, Scott J R. Excision and insertion of the conjugal transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. - PubMed
-
- Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. - PubMed
-
- Clewell D B, Flannagan S E. The conjugative transposons of gram-positive bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 369–393.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

