Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses
- PMID: 10871282
- PMCID: PMC2175135
- DOI: 10.1083/jcb.149.7.1419
Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses
Abstract
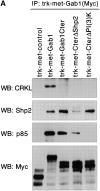
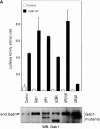
Gab1 is a substrate of the receptor tyrosine kinase c-Met and involved in c-Met-specific branching morphogenesis. It associates directly with c-Met via the c-Met-binding domain, which is not related to known phosphotyrosine-binding domains. In addition, Gab1 is engaged in a constitutive complex with the adaptor protein Grb2. We have now mapped the c-Met and Grb2 interaction sites using reverse yeast two-hybrid technology. The c-Met-binding site is localized to a 13-amino acid region unique to Gab1. Insertion of this site into the Gab1-related protein p97/Gab2 was sufficient to confer c-Met-binding activity. Association with Grb2 was mapped to two sites: a classical SH3-binding site (PXXP) and a novel Grb2 SH3 consensus-binding motif (PX(V/I)(D/N)RXXKP). To detect phosphorylation-dependent interactions of Gab1 with downstream substrates, we developed a modified yeast two-hybrid assay and identified PI(3)K, Shc, Shp2, and CRKL as interaction partners of Gab1. In a trk-met-Gab1-specific branching morphogenesis assay, association of Gab1 with Shp2, but not PI(3)K, CRKL, or Shc was essential to induce a biological response in MDCK cells. Overexpression of a Gab1 mutant deficient in Shp2 interaction could also block HGF/SF-induced activation of the MAPK pathway, suggesting that Shp2 is critical for c-Met/Gab1-specific signaling.
Figures





References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Astier A., Manie S.N., Law S.F., Canty T., Haghayghi N., Druker B.J., Salgia R., Golemis E.A., Freedman A.S. Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in human B-cellspotential relevance to neoplastic lymphohematopoietic cells. Leuk. Lymphoma. 1997;28:65–72. - PubMed
-
- Ausubel, F.M., R. Brent, R. Kingston, D. Moore, J. Seidmann, J. Smith, and K. Struhls. 1992. Curr. Prot. Mol. Biol.
-
- Bardelli A., Longati P., Gramaglia D., Stella M.C., Comoglio P.M. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene. 1997;15:3103–3111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

