Loss-of-function genetics in mammalian cells: the p53 tumor suppressor model
- PMID: 10871344
- PMCID: PMC102629
- DOI: 10.1093/nar/28.11.2234
Loss-of-function genetics in mammalian cells: the p53 tumor suppressor model
Abstract
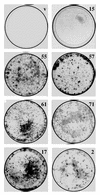
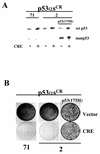
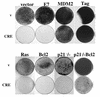
Using an improved system for the functional identification of active antisense fragments, we have isolated antisense fragments which inactivate the p53 tumour suppressor gene. These antisense fragments map in two small regions between nt 350 and 700 and nt 800 and 950 of the coding sequence. These antisense fragments appear to act by inhibition of p53 mRNA translation both in vivo and in vitro. Expression of these antisense fragments overcame the p53-induced growth arrest in a cell line which expresses a thermolabile mutant of p53 and extended the in vitro lifespan of primary mouse embryonic fibroblasts. Continued expression of the p53 antisense fragment contributed to immortalisation of primary mouse fibroblasts. Subsequent elimination of the antisense fragment in these immortalised cells led to restoration of p53 expression and growth arrest, indicating that immortal cells continuously require inactivation of p53. Expression of MDM2 or SV40 large T antigen, but not E7 nor oncogenic ras, overcomes the arrest induced by restoration of p53 expression. Functional inactivation of both p21 and bax (by overexpression of Bcl2), but not either alone, allowed some bypass of p53-induced growth arrest, indicating that multiple transcriptional targets of p53 may mediate its antiproliferative action. The ability to conditionally inactivate and subsequently restore normal gene function may be extremely valuable for genetic analysis of genes for which loss-of-function is involved in specific phenotypes.
Figures



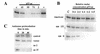
Similar articles
-
p16INK4A and p19ARF act in overlapping pathways in cellular immortalization.Nat Cell Biol. 2000 Mar;2(3):148-55. doi: 10.1038/35004020. Nat Cell Biol. 2000. PMID: 10707085
-
Transcriptional inactivation of p53, Bax, Bcl-2 and Mdm2 correlates with malignant transformation of the uterine cervix.Int J Biol Markers. 2005 Jan-Mar;20(1):18-27. Int J Biol Markers. 2005. PMID: 15832769
-
Experimental therapy of human prostate cancer by inhibiting MDM2 expression with novel mixed-backbone antisense oligonucleotides: in vitro and in vivo activities and mechanisms.Prostate. 2003 Feb 15;54(3):194-205. doi: 10.1002/pros.10187. Prostate. 2003. PMID: 12518324
-
Tissue and cell-specific expression of the p53-target genes: bax, fas, mdm2 and waf1/p21, before and following ionising irradiation in mice.Oncogene. 2000 Feb 3;19(5):649-60. doi: 10.1038/sj.onc.1203366. Oncogene. 2000. PMID: 10698510
-
The p53 tumor suppressor gene and gene product.Princess Takamatsu Symp. 1989;20:221-30. Princess Takamatsu Symp. 1989. PMID: 2488233 Review.
Cited by
-
A dual-color fluorescence-based platform to identify selective inhibitors of Akt signaling.PLoS One. 2008 Mar 19;3(3):e1823. doi: 10.1371/journal.pone.0001823. PLoS One. 2008. PMID: 18350159 Free PMC article.
-
MAP17, a ROS-dependent oncogene.Front Oncol. 2012 Sep 6;2:112. doi: 10.3389/fonc.2012.00112. eCollection 2012. Front Oncol. 2012. PMID: 22973555 Free PMC article.
-
Cellular senescence as a target in cancer control.J Aging Res. 2010 Dec 30;2011:725365. doi: 10.4061/2011/725365. J Aging Res. 2010. PMID: 21234095 Free PMC article.
-
Cell Cycle Regulatory Protein Expression in Multinucleated Giant Cells of Giant Cell Tumor of Bone: do They Proliferate?Pathol Oncol Res. 2021 May 3;27:643146. doi: 10.3389/pore.2021.643146. eCollection 2021. Pathol Oncol Res. 2021. PMID: 34257609 Free PMC article.
-
Avian reovirus p17 and σA act cooperatively to downregulate Akt by suppressing mTORC2 and CDK2/cyclin A2 and upregulating proteasome PSMB6.Sci Rep. 2017 Jul 12;7(1):5226. doi: 10.1038/s41598-017-05510-x. Sci Rep. 2017. PMID: 28701787 Free PMC article.
References
-
- Serrano M., Lin,A.W., McCurrach,M.E., Beach,D. and Lowe,S.W. (1997) Cell, 88, 593–602. - PubMed
-
- Xu H.J., Zhou,Y., Ji,W., Perng,G.S., Kruzelock,R., Kong,C.T., Bast,R.C., Mills,G.B., Li,J. and Hu,S.X. (1997) Oncogene, 15, 2589–2596. - PubMed
-
- Ko L.J. and Prives,C. (1996) Genes Dev., 10, 1054–1072. - PubMed
-
- Lane D.P. (1994) Br. Med. Bull., 50, 582–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

