Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA
- PMID: 10871351
- PMCID: PMC102637
- DOI: 10.1093/nar/28.11.e52
Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA
Abstract
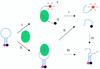
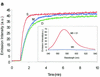
Traditional methods to assay enzymatic cleavage of DNA are discontinuous and time consuming. In contrast, recently developed fluorescence methods are continuous and convenient. However, no fluorescence method has been developed for single-stranded DNA digestion. Here we introduce a novel method, based on molecular beacons, to assay single-stranded DNA cleavage by single strand-specific nucleases. A molecular beacon, a hairpin-shaped DNA probe labeled with a fluorophore and a quencher, is used as the substrate and enzymatic cleavage leads to fluorescence enhancement in the molecular beacon. This method permits real time detection of DNA cleavage and makes it easy to characterize the activity of DNA nucleases and to study the steady-state cleavage reaction kinetics. The excellent sensitivity, reproducibility and convenience will enable molecular beacons to be widely useful for the study of single-stranded DNA cleaving reactions.
Figures





References
-
- Linn S.M. and Roberts,R.J. (1982) Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Linn S.M., Lloyd,R.S. and Roberts,R.J. (1993) Nucleases, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Mannual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- McLaughlin L.W., Benseler,F., Graeser,E., Piel,N. and Scholtissek,S. (1987) Biochemistry, 26, 7238–7245. - PubMed
-
- Alves J., Ruter,T., Geiger,R., Fliess,A., Maass,G. and Pingoud,A. (1989) Biochemistry, 28, 2678–2684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

