Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis
- PMID: 10871363
- PMCID: PMC102738
- DOI: 10.1093/nar/28.12.2315
Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis
Abstract
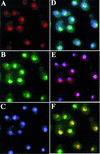
Ribosome biogenesis requires, in addition to rRNA molecules and ribosomal proteins, a multitude of trans-acting factors. Recently it has become clear that in the yeast Saccharomyces cerevisiae many RNA helicases of the DEAD-box and related families are involved in ribosome biogenesis. Here we show that the previously uncharacterised open reading frame YDL031w (renamed DBP10 for DEAD-box protein 10) encodes an essential putative RNA helicase that is required for accurate ribosome biogenesis. Genetic depletion of Dbp10p results in a deficit in 60S ribosomal subunits and an accumulation of half-mer polysomes. Furthermore, pulse-chase analyses of pre-rRNA processing reveal a strong delay in the maturation of 27SB pre-rRNA intermediates into 25S rRNA and 7S pre-rRNA. Northern blot analyses indicate that this delay leads to higher steady-state levels of 27SB species and reduced steady-state levels of 7S pre-rRNA and 25S/5.8S mature rRNAs, thus explaining the final deficit in 60S subunit and the formation of half-mer polysomes. Consistent with a direct role in ribosome biogenesis, Dbp10p was found to be located predominantly in the nucleolus.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

