Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803
- PMID: 10871367
- PMCID: PMC102721
- DOI: 10.1093/nar/28.12.2353
Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803
Abstract
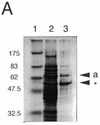
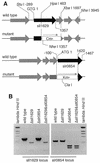
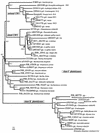
Photolyase is a DNA repair enzyme that reverses UV-induced photoproducts in DNA in a light-dependent manner. Recently, photolyase homologs were identified in higher eukaryotes. These homologs, termed crypto-chromes, function as blue light photoreceptors or regulators of circadian rhythm. In contrast, most bacteria have only a single photolyase or photolyase-like gene. Unlike other microbes, the chromosome of the cyanobacterium SYNECHOCYSTIS: sp. PCC6803 contains two ORFs (slr0854 and sll1629) with high similarities to photolyases. We have characterized both genes. The slr0854 gene product exhibited specific, light-dependent repair activity for a cyclo-butane pyrimidine dimer (CPD), whereas the sll1629 gene product lacks measurable affinity for DNA in vitro. Disruption of either slr0854 or sll1629 had little or no effect on the growth rate of the cyanobacterium. A mutant lacking the slr0854 gene showed severe UV sensitivity, in contrast to a mutant lacking sll1629. Phylogenetic analysis showed that sll1629 is more closely related to the cryptochromes than photolyases. We conclude that sll1629 is a bacterial cryptochrome. To our knowledge, this is the first description of a bacterial cryptochrome.
Figures






References
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis, American Society for Microbiology, Washington, DC.
-
- Sancar A. (1994) Biochemistry, 33, 2–9. - PubMed
-
- Todo T., Takemori,H., Ryo,H., Ihara,M., Matsunaga,T., Nikaido,O., Sato,K. and Nomura,T. (1993) Nature, 361, 371–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

