DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1
- PMID: 10871372
- PMCID: PMC102730
- DOI: 10.1093/nar/28.12.2389
DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1
Abstract
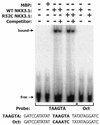
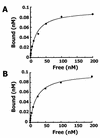
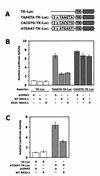
NKX3.1 is a member of the NK class of homeodomain proteins and is most closely related to Drosophila NK-3. NKX3.1 has predominantly prostate-specific expression in the adult human. Previous studies suggested that NKX3.1 exerts a growth-suppressive effect on prostatic epithelial cells and controls differentiated glandular functions. Using a binding site selection assay with recombinant NKX3.1 protein we identified a TAAGTA consensus binding sequence that has not been reported for any other NK class homeoprotein. By electromobility shift assay we demonstrated that NKX3.1 preferentially binds the TAAGTA sequence rather than the binding site for Nkx2.1 (CAAGTG) or Msx1 (TAATTG). Using mutated binding sites in competitive gel shift assays, we analyzed the nucleotides in the TAAGTA consensus sequence that are important for NKX3.1 binding. The consensus binding site of a naturally occurring polymorphic NKX3.1 protein with arginine replaced by cysteine at position 52 was identical to the wild-type binding sequence. The binding affinities of wild-type and polymorphic NKX3.1 for the TAAGTA consensus site were very similar, with values of 20 and 22 nM, respectively. Wild-type and polymorphic NKX3.1 specifically repressed transcription of luciferase from a reporter vector with three copies of the NKX3.1-binding site upstream from a thymidine kinase promoter. The data show that among NK family proteins NKX3.1 binds a novel DNA sequence and can behave as an in vitro transcriptional repressor.
Figures



Similar articles
-
Characterization of Nkx3.2 DNA binding specificity and its requirement for somitic chondrogenesis.J Biol Chem. 2003 Jul 25;278(30):27532-9. doi: 10.1074/jbc.M301461200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12746429
-
Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor.J Mol Biol. 2006 Jul 28;360(5):989-99. doi: 10.1016/j.jmb.2006.05.064. Epub 2006 Jun 15. J Mol Biol. 2006. PMID: 16814806
-
Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function.Cancer Res. 2002 May 1;62(9):2654-9. Cancer Res. 2002. PMID: 11980664
-
Regulating NKX3.1 stability and function: Post-translational modifications and structural determinants.Prostate. 2016 May;76(6):523-33. doi: 10.1002/pros.23144. Epub 2016 Feb 4. Prostate. 2016. PMID: 26841725 Review.
-
Mechanisms of prostate tumorigenesis: roles for transcription factors Nkx3.1 and Egr1.Ann N Y Acad Sci. 2005 Nov;1059:33-40. doi: 10.1196/annals.1339.018. Ann N Y Acad Sci. 2005. PMID: 16382041 Review.
Cited by
-
Structural and functional interactions of the prostate cancer suppressor protein NKX3.1 with topoisomerase I.Biochem J. 2013 Jul 1;453(1):125-36. doi: 10.1042/BJ20130012. Biochem J. 2013. PMID: 23557481 Free PMC article.
-
Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription.Mol Cell Biol. 2003 Oct;23(19):7019-29. doi: 10.1128/MCB.23.19.7019-7029.2003. Mol Cell Biol. 2003. PMID: 12972618 Free PMC article.
-
NKL homeobox gene activities in hematopoietic stem cells, T-cell development and T-cell leukemia.PLoS One. 2017 Feb 2;12(2):e0171164. doi: 10.1371/journal.pone.0171164. eCollection 2017. PLoS One. 2017. PMID: 28151996 Free PMC article.
-
Gene expression profiles in the PC-3 human prostate cancer cells induced by NKX3.1.Mol Biol Rep. 2010 Mar;37(3):1505-12. doi: 10.1007/s11033-009-9549-8. Epub 2009 May 22. Mol Biol Rep. 2010. PMID: 19462257
-
CRISPR/Cas9-Mediated Point Mutation in Nkx3.1 Prolongs Protein Half-Life and Reverses Effects Nkx3.1 Allelic Loss.Cancer Res. 2020 Nov 1;80(21):4805-4814. doi: 10.1158/0008-5472.CAN-20-1742. Epub 2020 Sep 17. Cancer Res. 2020. PMID: 32943441 Free PMC article.
References
-
- Gehring W.J., Affolter,M. and Burglin,T. (1994) Annu. Rev. Biochem., 63, 487–526. - PubMed
-
- He W.W., Sciavolino,P.J., Wing,J., Augustus,M., Hudson,P., Meissner,P.S., Curtis,R.T., Shell,B.K., Bostwick,D.G., Tindall,D.J. et al. (1997) Genomics, 43, 69–77. - PubMed
-
- Prescott J.L., Blok,L. and Tindall,D.J. (1998) Prostate, 35, 71–80. - PubMed
-
- Sciavolino P.J., Abrams,E.W., Yang,L., Austenberg,L.P., Shen,M.M. and Abate-Shen,C. (1997) Dev. Dyn., 209, 127–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

