Quantitative characterization of the interaction between purified human estrogen receptor alpha and DNA using fluorescence anisotropy
- PMID: 10871398
- PMCID: PMC102715
- DOI: 10.1093/nar/28.13.2494
Quantitative characterization of the interaction between purified human estrogen receptor alpha and DNA using fluorescence anisotropy
Abstract
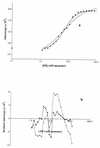
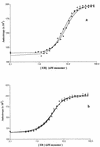
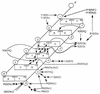
In an effort to better define the molecular mechanisms of the functional specificity of human estrogen receptor alpha, we have carried out equilibrium binding assays to study the interaction of the receptor with a palindromic estrogen response element derived from the vitellogenin ERE. These assays are based on the observation of the fluorescence anisotropy of a fluorescein moiety covalently bound to the target oligonucleotide. The low anisotropy value due to the fast tumbling of the free oligonucleotide in solution increases substantially upon binding the receptor to the labeled ERE. The quality of our data are sufficient to ascertain that the binding is clearly cooperative in nature, ruling out a simple monomer interaction and implicating a dimerization energetically coupled to DNA binding in the nanomolar range. The salt concentration dependence of the affinity reveals formation of high stoichiometry, low specificity complexes at low salt concentration. Increasing the KCl concentration above 200 mM leads to specific binding of ER dimer. We interpret the lack of temperature dependence of the apparent affinity as indicative of an entropy driven interaction. Finally, binding assays using fluorescent target EREs bearing mutations of each of the base pairs in the palindromic ERE half-site indicate that the energy of interaction between ER and its target is relatively evenly distributed throughout the site.
Figures




References
-
- McKenna N.J., Lanz,R.B. and O’Malley,B.W. (1999) Endocrin. Rev., 20, 321–344. - PubMed
-
- Murdoch F.E., Meier,D.A., Furlow,J.D., Grunwald,K.A.A. and Gorski,J. (1990) Biochemistry, 29, 8377–8385. - PubMed
-
- Murdoch F.E., Grunwald,K.A.A. and Gorski,J. (1991) Biochemistry, 30, 10838–10844. - PubMed
-
- Furlow J.D., Murdoch,F.E. and Gorski,J. (1993) J. Biol. Chem., 268, 12519–12525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

