Importance of discriminator base stacking interactions: molecular dynamics analysis of A73 microhelix(Ala) variants
- PMID: 10871402
- PMCID: PMC102696
- DOI: 10.1093/nar/28.13.2527
Importance of discriminator base stacking interactions: molecular dynamics analysis of A73 microhelix(Ala) variants
Abstract
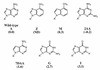
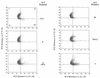
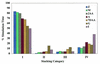
Transfer of alanine from Escherichia coli alanyl-tRNA synthetase (AlaRS) to RNA minihelices that mimic the amino acid acceptor stem of tRNA(Ala) has been shown, by analysis of variant minihelix aminoacylation activities, to involve a transition state sensitive to changes in the 'discriminator' base at position 73. Solution NMR has indicated that this single-stranded nucleotide is predominantly stacked onto G1 of the first base pair of the alanine acceptor stem helix. We report the activity of a new variant with the adenine at position 73 substituted by its non-polar isostere 4-methylindole (M). Despite lacking N7, this analog is well tolerated by AlaRS. Molecular dynamics (MD) simulations show that the M substitution improves position 73 base stacking over G1, as measured by a stacking lifetime analysis. Additional MD simulations of wild-type microhelix(Ala) and six variants reveal a positive correlation between N73 base stacking propensity over G1 and aminoacylation activity. For the two DeltaN7 variants simulated we found that the propensity to stack over G1 was similar to the analogous variants that contain N7 and we conclude that the decrease in aminoacylation efficiency observed upon deletion of N7 is likely due to loss of a direct stabilizing interaction with the synthetase.
Figures


References
-
- Hou Y.-M. (1997) Chem. Biol., 4, 93–96. - PubMed
-
- Musier-Forsyth K. (1998) J. Biochem. Mol. Biol., 31, 525–535.
-
- Musier-Forsyth K. and Schimmel,P. (1999) Acc. Chem. Res., 32, 368–375.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

