Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile
- PMID: 10878030
- PMCID: PMC86949
- DOI: 10.1128/JCM.38.7.2484-2487.2000
Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile
Abstract
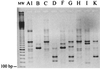
Clostridium difficile is now recognized as the major agent responsible for nosocomial diarrhea in adults. Among the genotyping methods available, arbitrarily primed PCR (AP-PCR), PCR-ribotyping, and pulsed-field gel electrophoresis (PFGE) have been widely used for investigating outbreaks of C. difficile infections. However, the comparative typing ability, reproducibility, discriminatory power, and efficiency of these methods have not been fully investigated. We compared the results of three methods-AP-PCR with three different primers (AP3, AP4, and AP5), PCR-ribotyping, and PFGE (with SmaI endonuclease)-to differentiate 99 strains of C. difficile that had been previously serogrouped. Typing abilities were 100% for PCR-ribotyping and AP-PCR with AP3 and 90% for PFGE, due to early DNA degradation in strains from serogroup G. Reproducibilities were 100% for PCR-ribotyping and PFGE but only 88% for AP-PCR with AP3, 67% for AP-PCR with AP4, and 33% for AP-PCR with AP5. Discriminatory power for unrelated strains was >0.95 for all the methods but was lower for PCR-ribotyping among serogroups D and C. PCR-based methods were easier and quicker to perform, but their fingerprints were more difficult to interpret than those of PFGE. We conclude that PCR-ribotyping offers the best combination of advantages as an initial typing tool for C. difficile.
Figures


References
-
- Barbut F, Corthier G, Charpak Y, Cerf M, Monteil H, Fosse T, Trevoux A, De Barbeyrac B, Boussougant Y, Tigaud S, Tytgat F, Sedallian A, Duborgel S, Collignon A, Le Guern M E, Bernasconi P, Petit J C. Prevalence and pathogenicity of Clostridium difficile in hospitalized patients. Arch Intern Med. 1996;156:1449–1454. - PubMed
-
- Barbut F, Mario N, Delmée M, Gozlan J, Petit J C. Genomic fingerprinting of Clostridium difficile isolates by using a random amplified polymorphic DNA (RAPD) assay. FEMS Microbiol Lett. 1993;114:161–166. - PubMed
-
- Bartlett J G. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about this organism. Clin Infect Dis. 1994;18(Suppl. 4):S265–S272. - PubMed
-
- Bidet P, Barbut F, Lalande V, Burghoffer B, Petit J C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. - PubMed
-
- Bowman R A, O'Neill G L, Riley T V. Non-radioactive restriction fragment length polymorphism (RFLP) typing of Clostridium difficile. FEMS Microbiol Lett. 1991;79:269–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

