Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2
- PMID: 10878032
- PMCID: PMC86951
- DOI: 10.1128/JCM.38.7.2494-2503.2000
Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2
Abstract
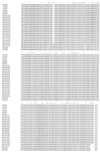
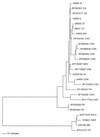
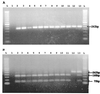
Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in swine. Increasing evidence indicates that a variant strain of porcine circovirus (PCV), designated type 2 PCV (PCV-2), is responsible for PMWS. To determine the extent of genetic heterogeneity of PCV-2 isolates, the complete genomes of six PCV-2 isolates from different regions of North America were amplified by PCR and sequenced. Sequence and phylogenetic analyses confirmed that two distinct genotypes of PCV exist: nonpathogenic genotype PCV-1 and PMWS-associated genotype PCV-2. However, within the PCV-2 genotype, several minor branches that have been identified appear to be associated with geographic origins. The genomic sequences of two French PCV-2 isolates diverge the most from those of other PCV-2 isolates and form a distinct branch. Other minor but distinguishable branches have also been identified for a Taiwan PCV-2 isolate and two of the Canadian PCV-2 isolates. All the U.S. PCV-2 isolates are closely related, but the Canadian isolates vary, to some extent, in their genomic sequences. The data from this study indicate that although the genome of PCV-2 is generally stable among different isolates, PCV-2 isolates from different geographic regions vary in their genomic sequences. This variation may have important implications for PCV-2 diagnosis and research. On the basis of genetic analyses of available PCV strains, a universal PCR-restriction fragment length polymorphism (PCR-RFLP) assay was developed to detect and differentiate between infections with PCV-1 and PCV-2. This PCR-RFLP assay should be useful for studying the pathogenesis of PCV-2, for detecting PCV-2 infection in pigs from different geographic regions, and for screening donor pigs for use in xenotransplantation.
Figures


References
-
- Allan G M, McNeilly F, Cassidy J P, Reilly G A, Adair B, Ellis W A, McNulty M S. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. - PubMed
-
- Allan G M, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark E G, Harding J, Espuna E, Botner A, Charreyre C. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 1998;142:467–468. - PubMed
-
- Allan G M, McNeilly F, Kennedy S, Daft B, Clarke E G, Ellis J A, Haines D M, Meehan B M, Adair B M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. - PubMed
-
- Allan G M, McNeilly F, Meehan B M, Kennedy S, Mackie D P, Ellis J A, Clark E G, Espuna E, Saubi N, Riera P, Botner A, Charreyre C E. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol. 1999;66:115–123. - PubMed
-
- Allan G M, Kennedy S, McNeilly F, Foster J C, Ellis J A, Krakowka S J, Meehan B M, Adair B M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol. 1999;121:1–11. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

