Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation
- PMID: 10878039
- PMCID: PMC86962
- DOI: 10.1128/JCM.38.7.2536-2542.2000
Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation
Abstract
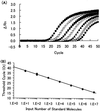
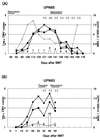
The purpose of this study was to assess the usefulness of real-time automated PCR as a quantitative, highly reproducible, and sensitive method to detect cytomegalovirus (CMV) DNA in blood specimens. Intra- and interassay precision rates were 0.89% (small number of copies [L]), 1.43% (middle number of copies [M]), and 1.12% (high number of copies [H]), and 4.46% (L), 1.51% (M), and 2.28% (H), respectively. The linearity of this assay was obtained between 10 and 10(7) copies/well, with a minimum detection limit of 20 copies/well. Specimens from 55 of 70 healthy subjects were found to be positive for CMV antibody, but CMV DNA was not detected in any of them. In the qualitative assessment of each specimen, the results of the CMV antigenemia assay and those of the real-time PCR assay agreed in 80% (plasma specimens), 79% (all nucleated cells), and 86% (blood) of the cases examined. For eight patients diagnosed as having CMV infection or disease, no sample was positive in the antigenemia assay earlier than in the real-time PCR assay. Furthermore, the results of this assay could be obtained within 8 h. We concluded that the real-time PCR assay is useful for rapid diagnosis of CMV infection and monitoring of clinical courses.
Figures

References
-
- Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Chee M S, Hutchison C A D, Kouzarides T, Martignetti J A, Preddie E. The DNA sequence of the human cytomegalovirus genome. DNA Sequences. 1991;2:1–12. - PubMed
-
- Boeckh M, Gallez-Hawkins G M, Myerson D, Zaia J A, Bowden R A. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation. 1997;64:108–113. - PubMed
-
- Boeckh M, Gooley T A, Myerson D, Cunningham T, Schoch G, Bowden R A. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood. 1996;88:4063–4071. - PubMed
-
- Goodrich J M, Mori M, Gleaves C A, Du Mond C, Cays M, Ebeling D F, Buhles W C, DeArmond B, Meyers J D. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplantation. N Engl J Med. 1991;325:1601–1607. - PubMed
-
- Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

