Circadian clock-protein expression in cyanobacteria: rhythms and phase setting
- PMID: 10880447
- PMCID: PMC313937
- DOI: 10.1093/emboj/19.13.3349
Circadian clock-protein expression in cyanobacteria: rhythms and phase setting
Abstract
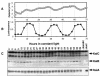
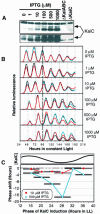
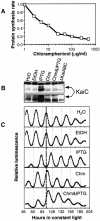
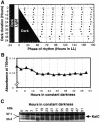
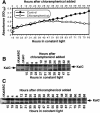
The cyanobacterial gene cluster kaiABC encodes three essential circadian clock proteins: KaiA, KaiB and KaiC. The KaiB and KaiC protein levels are robustly rhythmical, whereas the KaiA protein abundance undergoes little if any circadian oscillation in constant light. The level of the KaiC protein is crucial for correct functioning of the clock because induction of the protein at phases when the protein level is normally low elicits phase resetting. Titration of the effects of the inducer upon phase resetting versus KaiC level shows a direct correlation between induction of the KaiC protein within the physiological range and significant phase shifting. The protein synthesis inhibitor chloramphenicol prevents the induction of KaiC and blocks phase shifting. When the metabolism is repressed by either translational inhibition or constant darkness, the rhythm of KaiC abundance persists; therefore, clock protein expression has a preferred status under a variety of conditions. These data indicate that rhythmic expression of KaiC appears to be a crucial component of clock precession in cyanobacteria.
Figures

References
-
- Aronson B., Johnson,K., Loros,J.J. and Dunlap,J.C. (1994) Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science, 263, 1578–1584. - PubMed
-
- Darlington T.K., Wager-Smith,K., Ceriani,M.F., Staknis,D., Gekakis,N., Steeves,T.D.L., Weitz,C.J., Takahashi,J.S. and Kay,S.A. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science, 280, 1599–1603. - PubMed
-
- Doolittle W.F. (1979) The cyanobacterial genome, its expression and the control of that expression. Adv. Microb. Physiol., 20, 1–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

