The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability
- PMID: 10880449
- PMCID: PMC313939
- DOI: 10.1093/emboj/19.13.3366
The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability
Abstract
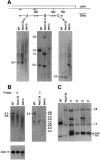
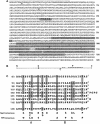
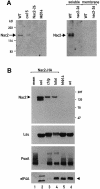
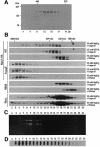
The psbD mRNA, which encodes the D2 reaction center polypeptide of photosystem II, is one of the most abundant chloroplast mRNAs. We have used genomic complementation to isolate the nuclear Nac2 gene, which is required for the stable accumulation of the psbD mRNA in Chlamydomonas reinhardtii. Nac2 encodes a hydrophilic polypeptide of 1385 amino acids with nine tetratricopeptide-like repeats (TPRs) in its C-terminal half. Cell fractionation studies indicate that the Nac2 protein is localized in the stromal compartment of the chloroplast. It is part of a high molecular weight complex that is associated with non-polysomal RNA. Change of a conserved alanine residue of the fourth TPR motif by site-directed mutagenesis leads to aggregation of Nac2 protein and completely abrogates its function, indicating that this TPR is important for proper folding of the protein and for psbD mRNA stability, processing and/or translation.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

