HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins
- PMID: 10880450
- PMCID: PMC313940
- DOI: 10.1093/emboj/19.13.3377
HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins
Abstract
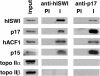
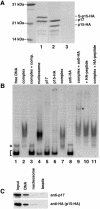
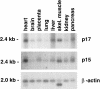
Chromatin remodelling complexes containing the nucleosome-dependent ATPase ISWI were first isolated from Drosophila embryos (NURF, CHRAC and ACF). ISWI was the only common component reported in these complexes. Our purification of human CHRAC (HuCHRAC) shows that ISWI chromatin remodelling complexes can have a conserved subunit composition in completely different cell types, suggesting a conserved function of ISWI. We show that the human homologues of two novel putative histone-fold proteins in Drosophila CHRAC are present in HuCHRAC. The two human histone-fold proteins form a stable complex that binds naked DNA but not nucleosomes. HuCHRAC also contains human ACF1 (hACF1), the homologue of Acf1, a subunit of Drosophila ACF. The N-terminus of mouse ACF1 was reported as a heterochromatin-targeting domain. hACF1 is a member of a family of proteins with a related domain structure that all may target heterochromatin. We discuss a possible function for HuCHRAC in heterochromatin dynamics. HuCHRAC does not contain topoisomerase II, which was reported earlier as a subunit of Drosophila CHRAC.
Figures




Similar articles
-
Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling.EMBO J. 2001 Jul 16;20(14):3781-8. doi: 10.1093/emboj/20.14.3781. EMBO J. 2001. PMID: 11447119 Free PMC article.
-
Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC).EMBO J. 2000 Jun 15;19(12):3049-59. doi: 10.1093/emboj/19.12.3049. EMBO J. 2000. PMID: 10856248 Free PMC article.
-
The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF.Mol Cell. 2004 Jan 30;13(2):265-77. doi: 10.1016/s1097-2765(03)00523-9. Mol Cell. 2004. PMID: 14759371
-
SWItched-on mobility.Curr Biol. 1999 Oct 7;9(19):R742-6. doi: 10.1016/s0960-9822(99)80473-4. Curr Biol. 1999. PMID: 10530996 Review.
-
Regulation of ISWI chromatin remodelling activity.Chromosoma. 2014 Mar;123(1-2):91-102. doi: 10.1007/s00412-013-0447-4. Epub 2014 Jan 12. Chromosoma. 2014. PMID: 24414837 Review.
Cited by
-
Chromatin Remodelers: From Function to Dysfunction.Genes (Basel). 2015 Jun 12;6(2):299-324. doi: 10.3390/genes6020299. Genes (Basel). 2015. PMID: 26075616 Free PMC article. Review.
-
Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2.Mol Cell Biol. 2004 Nov;24(22):10047-57. doi: 10.1128/MCB.24.22.10047-10057.2004. Mol Cell Biol. 2004. PMID: 15509805 Free PMC article.
-
ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy.Adv Cancer Res. 2014;121:183-233. doi: 10.1016/B978-0-12-800249-0.00005-6. Adv Cancer Res. 2014. PMID: 24889532 Free PMC article. Review.
-
ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts.EMBO J. 2004 Oct 13;23(20):4029-39. doi: 10.1038/sj.emboj.7600382. Epub 2004 Sep 30. EMBO J. 2004. PMID: 15457208 Free PMC article.
-
Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates.Mol Cell Biol. 2001 Mar;21(6):2098-106. doi: 10.1128/MCB.21.6.2098-2106.2001. Mol Cell Biol. 2001. PMID: 11238944 Free PMC article.
References
-
- Aasland R., Gibson,T. and Stewart,A.F. (1995) The PHD-finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
-
- Aihara T., Miyoshi,Y., Koyama,K., Suzuki,M., Takahashi,E., Monden,M. and Nakamura,Y. (1998) Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet. Cell Genet., 81, 191–193. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

